|
September
1st
- 5 ml cultures of K1319003 and K1319004
- plasmid prep
| Plasmid | DNA [ng/µl]
|
| J23101.K516032 pSB1K3 | 23.5
|
| J23115.K516032 pSB1K3 | 20.5
|
| J04450 pSB1A3 | 57.5
|
| J04450 pSB1K1 | 63.5
|
- over night cultures of K131026 in DH5α and NEB
2nd
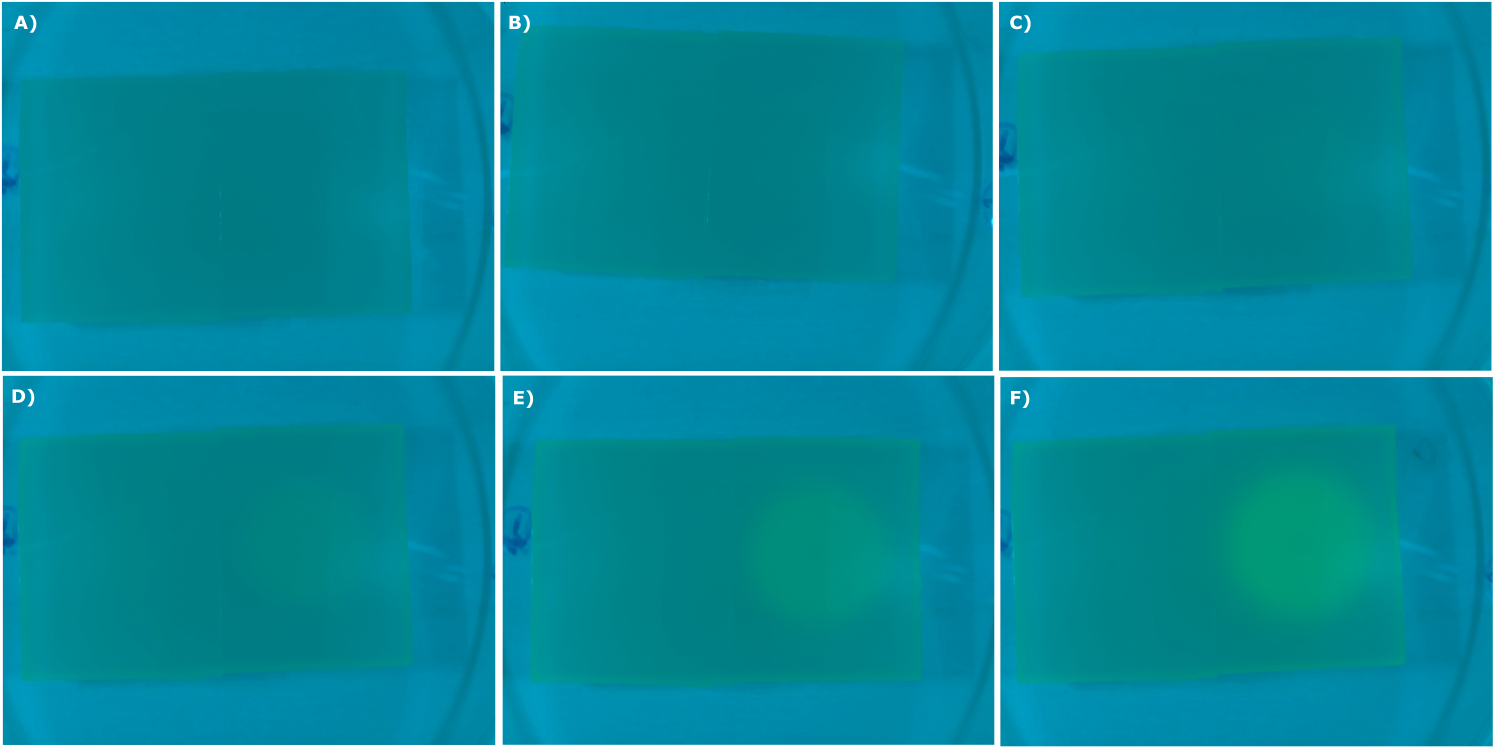
- made chips with K131026 in DH5α and NEB. Images were taken every 30 min with our own device
- gel purification of vector backbones
- sent to sequencing:
- K1318003
- K1319004
- J23101.K516032
- J23115.K516032
3rd
- prepared 50 mL LB+antibiotic overnight-cultures of pSBX-vectors that were sent in by team Heidelberg.
4th
- In the morning, at 10:15, we inoculated the precultures for the interlab study experiment.
- prepared cryo stocks of the pSBX-carrying E. coli from the overnight cultures. He also purified each pSBX-vector, eluting with 15+30 µL water, and resulting in the following DNA concentrations:
| vector | concentration [ng/µL]
|
| pSBX1A3 | 111
|
| pSBX4A5 | 14.1
|
| pSBX1C3 | 31
|
| pSB4C5 | 98.5
|
| pSBX1K3 | 18
|
| pSBX4K5 | 30
|
| pSBX1T3 | 39
|
| constitutive expression plasmid | 73
|
- PCRs for Gibson assembly of K1319003 into pET17. Duplicates of 25 µL reaction volume (12.5 µL Q5 2x Master Mix, 1.25 µL per primer, 2 µL template)
| PCR tube # | components
|
| 1 and 2 | pET17 + pET17_Gal3_Gib_F + pET17_Gal3_Gib_R
|
| 3 and 4 | K1319003 + K1319003_Gib_F + K1319003_Gib_R
|
The PCR conditions:
| step | temperature [°C] | duration
|
| denature | 98 | 30", 98 °C for 10", 55 °C for 30", 72 °C for 2'15"
|
| denature | 98 | 10"
|
| anneal | 50 (insert) 55 (backbone) | 30"
|
| elongate | 72 | 0'30" (insert) 2'15" (backbone)
|
| elongate | 72 | 2"
|
| store | 8 | indefinite
|
- Finally, we did the Gibson assembly and a heat shock transformation into NEB10β cells.
- At 10:15, we inoculated the primary cultures of the interlab study experiment and began with regular fluorescence measurements.
5th
- made master plates of yesterday's transformed cells.
6th
- made precultures of 3 clones from each prepared master palte and inoculated precultures for OD/F measurements as well as chip production on the 7th.
7th
- made cryos stocks of the precultures
- made chips with K131026 in DH5α and NEB and B0015 in NEB. Images were taken every 30 min with our own device
- purification of the following plasmids:
| plasmid | strain | resistance | vector | # of clone picked | concentration [ng/µl]
|
| K1319000 in I20260 | NEB10ß | K | pSB3K3 | 1 |
|
| K1319000 in I20260 | NEB10ß | K | pSB3K3 | 3 |
|
| K1319000 in I20260 | NEB10ß | K | pSB3K3 | 5 |
|
| K1319001 in I20260 | NEB10ß | K | pSB3K3 | 1 |
|
| K1319001 in I20260 | NEB10ß | K | pSB3K3 | 5 |
|
| K1319001 in I20260 | NEB10ß | K | pSB3K3 | 6 |
|
| K1319002 in I20260 | NEB10ß | K | pSB3K3 | 1 |
|
| K1319002 in I20260 | NEB10ß | K | pSB3K3 | 5 |
|
| K1319002 in I20260 | NEB10ß | K | pSB3K3 | 6 |
|
| K1319001_GFP Fusion in I20260 | NEB10ß | K | pSB3K3 | 4 |
|
| K1319001_GFP Fusion in I20260 | NEB10ß | K | pSB3K3 | 5 |
|
| K1319001_GFP Fusion in I20260 | NEB10ß | K | pSB3K3 | 6 |
|
| K1319002_GFP Fusion in I20260 | NEB10ß | K | pSB3K3 | 3 |
|
| K1319002_GFP Fusion in I20260 | NEB10ß | K | pSB3K3 | 4 |
|
| K1319002_GFP Fusion in I20260 | NEB10ß | K | pSB3K3 | 5 |
|
| His-SNAP-YFP-K1319003 | NEB10ß | A | pET17 | 3 |
|
| His-SNAP-YFP-K1319003 | NEB10ß | A | pET17 | 4 |
|
| His-SNAP-YFP-K1319003 | NEB10ß | A | pET17 | 6 |
|
Elution was performed twice with 15 µL of nuclease free water each time.
9th
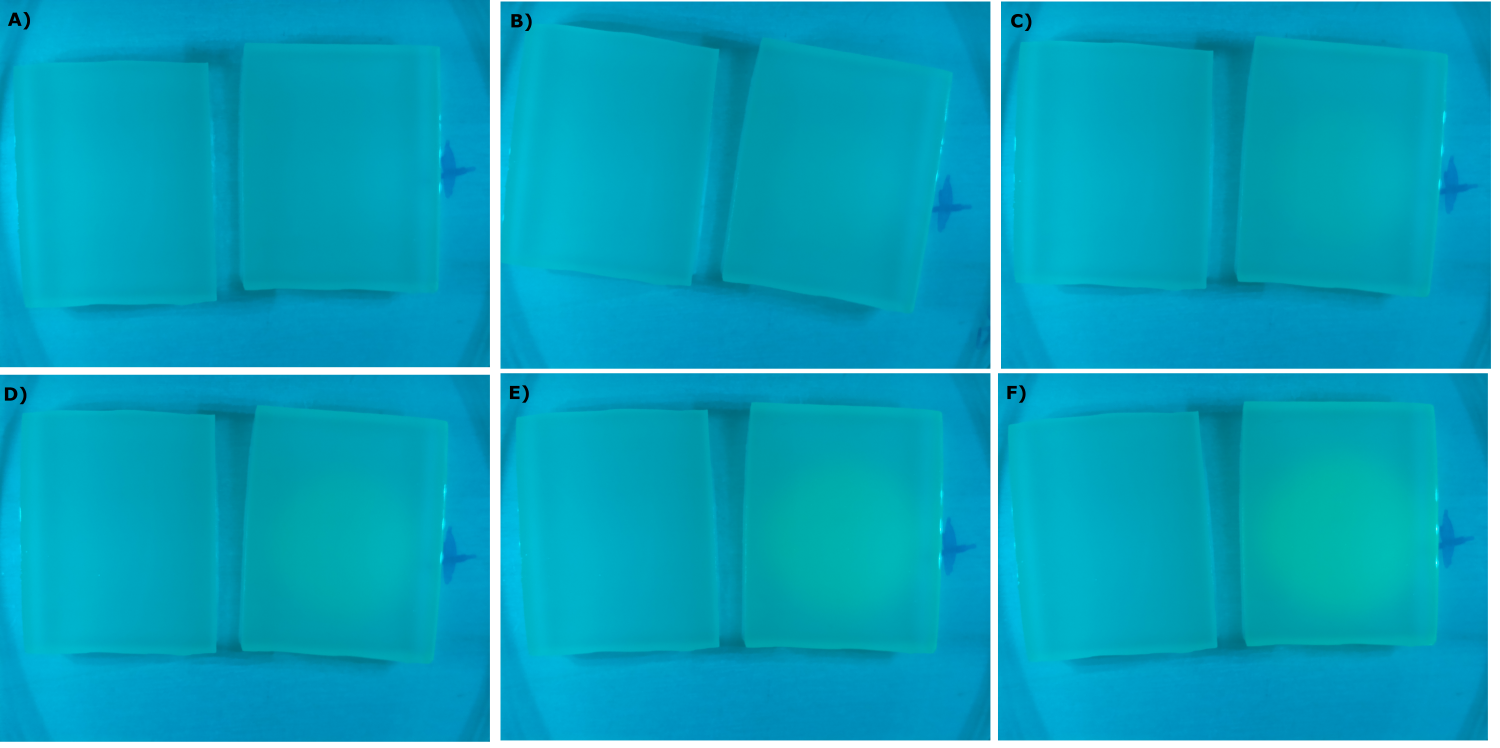
- made chips with K131026 in DH5α and NEB and without cells. Images were taken every 30 min with our own device
10th
- SDS page of REACh constructs after Gibson
- plasmid prep of Gal3 YFP
- #3: 20 ng/µl
- #4: 21.5 ng/µl
- #6: 15.9 ng/µl
15th
analyze the sequencing data from the clones of GFP_Reach 1, GFP_Reach 2 and K1319008.
GFP_Reach 2 clone #3 and #5 were fine, including the Leu to Ile mutation.
GFP_Reach 1 clone #4 and #5 were fine and did not contain the Leu to Ile mutation. Clone #6 was fine but contained the Leu to Ile mutation from the Reach 1 quick change mutations.
For future experiments, we will use the GFP_Reach 1 clone #4 and the GFP_Reach 2 clone #4.
Transformation of GFP_Reach 1 clone #3 and GFP_Reach 2 clones #3 and #5 were performed together with the TEV protease to create two plasmid construct.
The GFP_Reach 1 and GFP_Reach 2 constructs were also restricted and ligated into the pSB1C3 vector from the pSB3K3 vector.
- over night cultures of K131026 in DH5α and NEB
16th
- made master plates of the transformation from the day before.
- Also PCRs were made from pSBXA3, I20260 and K131900 for a Gibson assembly. The PCRs were checked with a gel electrophoresis.
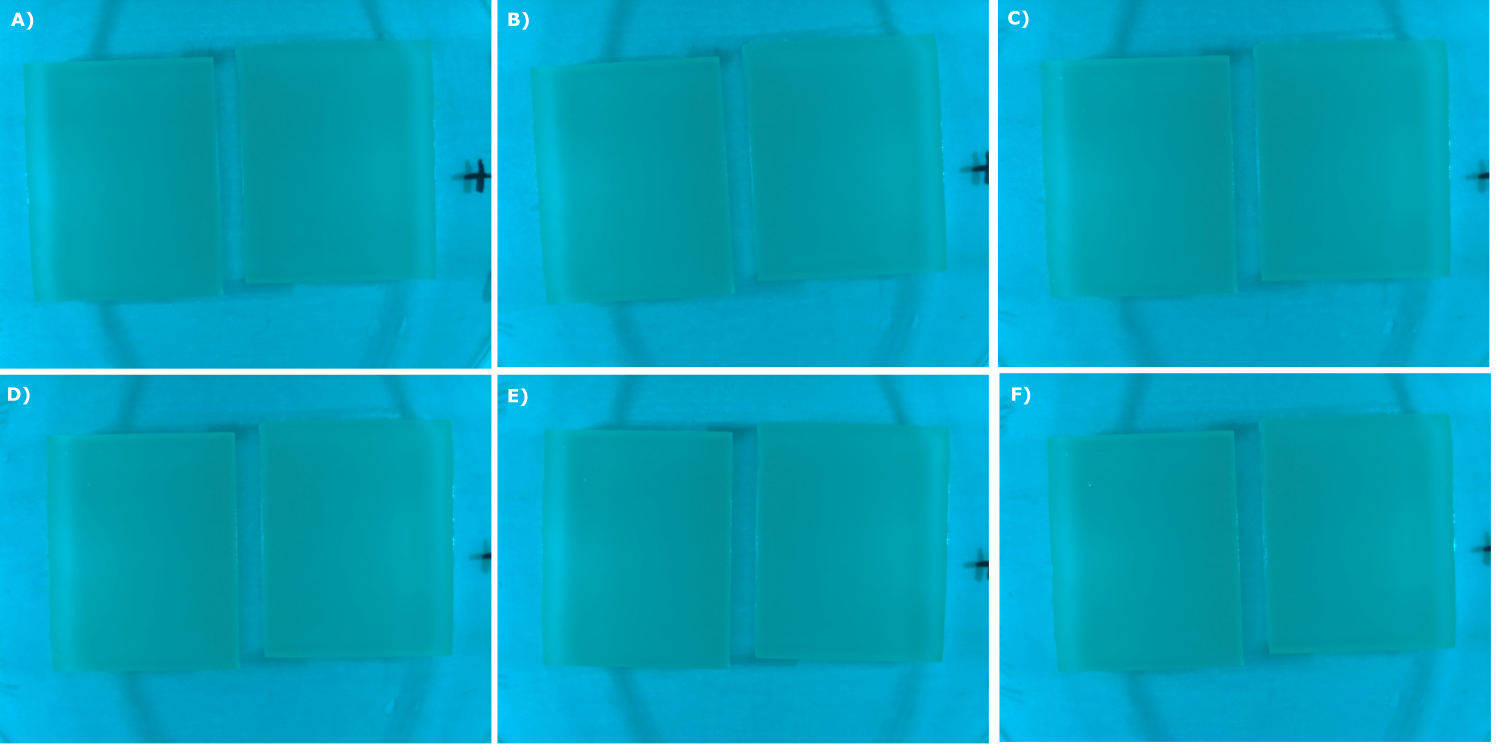
- made chips with K131026 in DH5α and NEB. Images were taken every 30 min with our own device
17th
- prepped and autoclaved 33 500 mL shake flasks.
18th
- SDS page of REACh constructs with TEV and IPTG
- over night cultures of K131026, B0015, K1319013, K1319014, K1319013 + K1319008 and K1319014 + K1319008 all in BL21
- tested Pseudomonas fluoresence if they are suitable for a growth experiment that is planned for our collaboration with the NEAnderLab next week. Therefore, she filled 2 500 mL flasks with 30 mL LB Pseudomonas-F medium, and inoculated each one with 1 mL culture medium of the overnight preculture. Flasks were inoculated at 30 °C at 250 rpm. However, after 5 hours no exponential growth could be shown (s. plot below). Thus, it was decided to use a E. coli K12 derivate strain in TB medium instead, and 30 mL of TB medium in a 500 mL flask were inoculated with E. coli DH5alpha cells and incubated at 37 °C at 300 rpm over night. According to the DSMZ E . coli K12 strain derivates, such as DH5alpha, are adequate for the kind of school experiment we are planning with the NEAnderLab.
19th
- made flask cultures of K1319013, K1319013 + K1319008, K1319013 + K1319008 + iPTG, K1319014, K1319014 + K1319008, K1319014 + K1319008 + iPTG, B0015 (negative control) and I20260 (positive control). iPTG was added at an OD of ~0.5. Inoculation was done via precultures in 500 ml shake flasks (50 ml filling volume). Media was always LB. Cultivation was done at 37 °C and 300 rpm. The starting OD was aimed to be 0.1. Inoculation occured directly from the precultures. Samples were taken every hour and checked for OD and fluorescence using a spectrophotometer and plate reader, respectively.
- did plasmid preparation from the cultures of the day before (K1319013, K1319013 + K1319008, K1319013 + K1319008 + iPTG, K1319014, K1319014 + K1319008, K1319014 + K1319008 + iPTG, B0015 and I20260). The plasmid were then be cut with EcoRI and PstI, and the results were be put on an agarose gel in order to perform a restriction test. Also plasmids of K1319013 and K1319014 will be cut with EcoRi and SpeI. K1319008 will be cut with XbaI and PstI. These will then be ligated together and then ligated into a pSB1A3 vector via the 3A assembly (vector cut with EcoRI and PstI). These constructs will be transformed into BL21 (and NEB as a backup). The created construts will be known as K1319018 (K1319013.K1319008) and K1319019 (K1319014.K1319008).
- made precultures of the master plates from the day before (K1319008, K1319013, K1319015 and pSBX1A3 with Gal3).
- also inoculated 4 cultures for the further testing of the OD/F device (the F part). The cultures are 2 shake flasks of I20260 and 2 shake flasks of B0015.
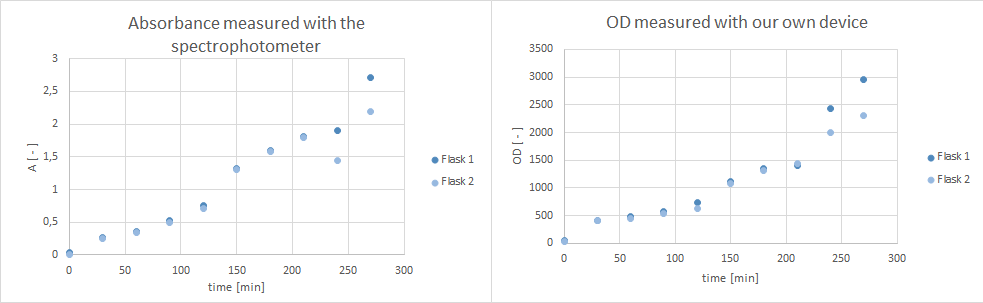
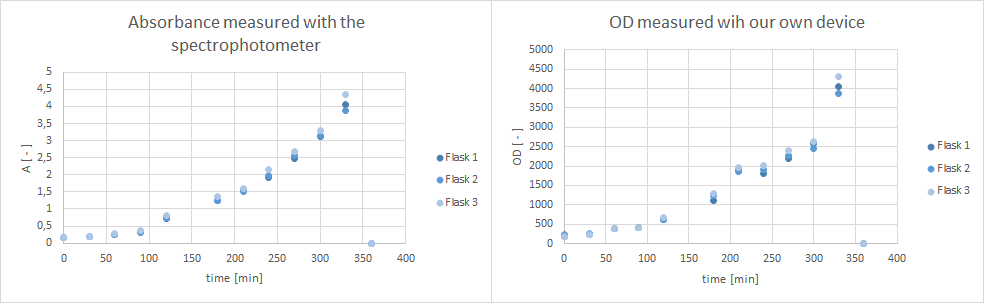
- Furthermore, did a growth experiment with DH5alpha for the NEAnderLab school experiment. 3 500 mL shake flasks were filled with 50 mL TB medium, and inoculated to an OD of 1.5 with the overnight preculture. Samples were taken every 30 minutes and tested for OD using our own device as well as the spectrophotometer. The resulting growth curve is shown below. we concluded that the growth was fast enough for these growth conditions to be used for the school experiment on the 24th.
- made chips with K1319013 + K1319008, K1319014 + K1319008, K1319013, K1319014, B0015 and K131026. Images were taken every 30 minutes with our own device.
- tested our OD/F device with a dilution test. Samples were checked with the spectrophotometer (OD), our OD/F device (fluorescence) and platereader (fluorescence).
- inoculated a culture of K1319008, B0015 as well as I20260 to check whether the results from our construct are from a wrongly done Gibson assembly with a still functioning superfolded GFP (the TEV protease was inserted in a backbone that formely contained superfolded GFP.)
20th
- SDS page of REACh constructs with TEV protease and induced by IPTG
22nd
- we poured several Pseudomonas-F agar plates with 0, 150 and 300 µg/L for the NEAnderLab school experiment. She also autoclaved 12 500 mL shake flasks, partly to be used for the school collaboration on Wednesday.
26th
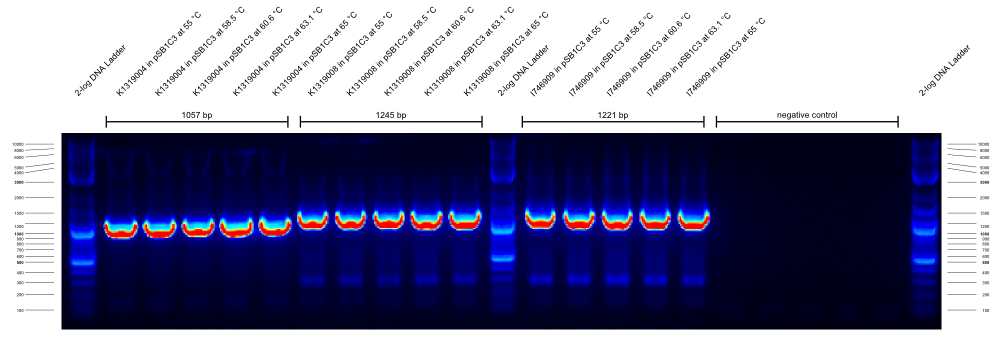
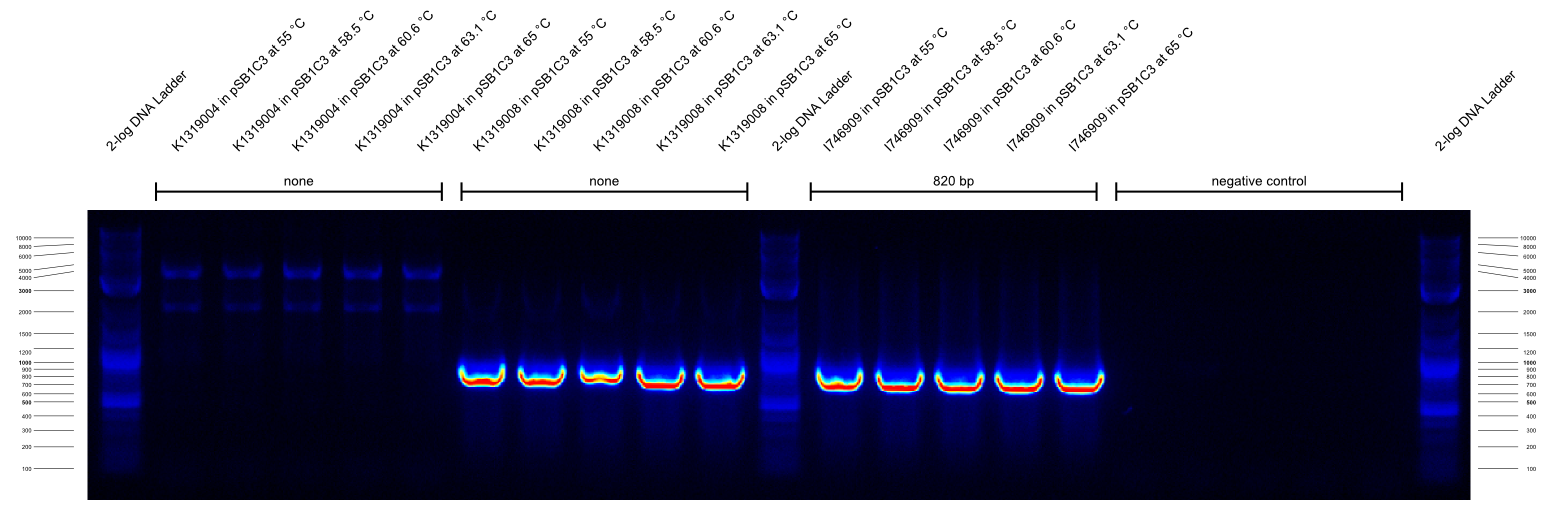
- We did a check PCR on several cryo cultures. All samples with G00100_Alternative+K1319004_check_R combinations resulted in a strong band at ~2300 bp that we cannot explain. All G00100_Alternative+K1319004_check_R combinations resulted in a strong band at 900 bp that we cannot explain either. We concluded that the annealing temperatures were wrong and favored unspecific products. Therefore, we decided to do a gradient PCR to find out the optimal annealing temperatures for our new primers.
- Gradient PCR to test new primer:
did gradient PCR with these new primers:
| name | sequence
|
| G00100_Alternative | GTGCCACCTGACGTCTAAGAAACCATTATTATC
|
| G00101_Alternative | ATTACCGCCTTTGAGTGAGCTGATACCGCTCG
|
| K1319004_check_R | ACGGAATTTCAGTTTCTGCGGGAACGGCGG
|
| I746909_check_R | ATCTTTAGACAGAACGCTTTGCGTGCTCAG
|
Three PCRs with different primer combinations were run. In all of them the templates were K1319004 in pSB1C3, K1319008 in pSB1C3 and I746909 in pSB1C3.
The first gradient PCR tested the G00100_Alternative + G00101_Alternative combination:
| primer_F | primer_R | template | expected length | best annealing temperature
|
| G00100_Alternative | G00101_Alternative | K1319004 in pSB1C3 | 1057 | ???
|
| G00100_Alternative | G00101_Alternative | K1319008 in pSB1C3 | 1245 | ???
|
| G00100_Alternative | G00101_Alternative | I746909 in pSB1C3 | 1221 | ???
|
| G00100_Alternative | G00101_Alternative | water | --- | ???
|
The second gradient PCR tested the G00100_Alternative + I746916_check_R combination:
| primer_F | primer_R | template | expected length | best annealing temperature
|
| G00100_Alternative | I746916_check_R | K1319004 in pSB1C3 | none | ???
|
| G00100_Alternative | I746916_check_R | K1319008 in pSB1C3 | none | ???
|
| G00100_Alternative | I746916_check_R | I746909 in pSB1C3 | 820 | ???
|
| G00100_Alternative | I746916_check_R | water | --- | ???
|
The third gradient PCR tested the G00100_Alternative + K1319004_check_R combination:
| primer_F | primer_R | template | expected length | best annealing temperature
|
| G00100_Alternative | K1319004_check_R | K1319004 in pSB1C3 | 541 | ???
|
| G00100_Alternative | K1319004_check_R | K1319008 in pSB1C3 | 502 | ???
|
| G00100_Alternative | K1319004_check_R | I746909 in pSB1C3 | none | ???
|
| G00100_Alternative | K1319004_check_R | water | --- | ???
|
The results of these three PCRs are:
- KAPA2G Fast ReadyMix worked well
- all three primers work well at >65 °C annealing temperature
- K1319008 template contained I746916 instead of the intended K1319004 ORF
It was concluded that a similar check PCR with 65 °C annealing temperature will be done on all plasmids and cryos of K1319008.
27th
- First we transformed K1319001, K1319002, K1319003 and K1319004 (all in pSB1C3) into NEB10β cells. He tested the PCR machine for semi-automated heat-shocking by splitting the 50 µL cells with the plasmid into 2x 25 µL. All 100 µL were plated for all construct/machine combinations.
- transformed several constructs into chemically competent BL21(DE3) cells.
- we did colony-PCR on all plasmids, cryos and colonies that should contain the K1319004 sequence.
- we also made check a PCR on galectin-constructs:
| label | primer_F | primer_R | expected length | result
|
| Gal3 in pSBX1A3 #1 | G00100_Alternative | K1319003_R | 1684 | ???
|
| Gal3 in pSBX1A3 #2 | G00100_Alternative | K1319003_R | 1684 | ???
|
| Gal3 in pSBX1A3 #3 | G00100_Alternative | K1319003_R | 1684 | ???
|
| Gal3 YFP #3 | pETGal3_seq_F | K1319003_R | 867 | ???
|
| Gal3 YFP #3 | pETGal3_seq_F | K1319003_R | 867 | ???
|
| Gal3 YFP pet17 AmpR | pETGal3_seq_F | K1319003_R | 867 or none | ???
|
| pET17 Gal3 #1 | pETGal3_seq_F | K1319003_R | none | ???
|
| K1319003 in pSB1C3 | G00100_Alternative | K1319003_R | 930 | ???
|
28th
- made a restriction of BioBrick K1319020 and vector pSB1C3 with restriction enzymes EcoRI and PstI. Then we ligated the restricted parts and made a transformation using E. coli NEB 10ß cells.
29th
- made cryo cultures and plasmid preparation of K1319010, K1319011, K1319012, K1319021 and K1319042. We determined the contentration of plasmids and made did a restriction digest of K1319010, K1319011, K1319012, pSB1C3, K1319021, K1319013 and K1319014, followed by a ligation in K1319010.pSB1C3, K1319011.pSB1C3, K1319012.pSB1C3, K1319021.K1319013.pSB1A3 and K1319021.K1319013.pSB1A3. All constructs were transformed into E. coli NEB 10ß.
- prepared 3 500 mL flasks with 30 mL LB medium which were inoculated with a Pseudomonas putida strain. The cells were cultured over night at 28 °C and ~300 rpm. The cultures are supposed to be used to test our OD device.
30th
- Sequencing samples were sent in for K1319020 clone #2, 3 & 5 (in pSB1C3), K1319017 clone #1 (in pSB1C3), K1319010 clone #2 (in pSB3K3), K1319011 clone #1 (in pSB3K3), K1319012 clone #2 (in pSB3K3), K1319013 clone #1 (in pSB1C3), K1319014 clone #1 (in pSB1C3), K1319001 (in pSB1C3) and K1319002 (in pSB1C3).
- A plasmid prep of K1319013 and K1319014 was run.
- A Gibson assembly with the K1319015 from the I20260 backbone and the K1319000 insert, forming K3139015, was conducted. The product was subsequently transformed into NEB10β cells.
- The pSB1C3 plasmid backbones were amplified via PCR and purified.
- Colony-PCRs of K1319008 and K1319012 master plates were made to confirm the colony's identity. Subsequently, pre-cultures were inoculated.
- A transformation of K1319010 and K1319010 in pSB1C3 was conducted.
- Another plasmid prep of K1319010 clone #2, K1319011 clone #1, K1319012 clone #2 (all in pSB3K3), K1319013 clone #4, K1319014 #3, K139020 #2, 3, 5 (all in pSB1C3) was run.
- The OD device was tested with a dilution series of a Pseudomonas putida culture.
|
 "
"