Team:Aachen/Notebook/Wetlab/August
From 2014.igem.org
(Difference between revisions)
(→5th) |
(→6th) |
||
| (6 intermediate revisions not shown) | |||
| Line 93: | Line 93: | ||
== 4th == | == 4th == | ||
| - | * made cryo stocks of K1319042 and K131026 in NEB/BL21/ | + | * made cryo stocks of K1319042 and K131026 in NEB/BL21/DH5α, I746909 in BL21 and pET17-His-SNAP-YFP-Gal3 in ''E. coli'' rosetta (DE3), respectively. |
* made plasmid prep, most of them using 1.5 mL culture medium, and eluted with 1x 50 µL of ddH{{sub|2}}O. The resulting DNA concentrations are shown below. | * made plasmid prep, most of them using 1.5 mL culture medium, and eluted with 1x 50 µL of ddH{{sub|2}}O. The resulting DNA concentrations are shown below. | ||
| Line 106: | Line 106: | ||
| I746909 BL21 #3 || 49 | | I746909 BL21 #3 || 49 | ||
|- | |- | ||
| - | | K1319042 | + | | K1319042 DH5α || 60 |
|- | |- | ||
| - | | K131026 | + | | K131026 DH5α || 150 |
|- | |- | ||
| pET17-Gal3 #1 || 30.5 | | pET17-Gal3 #1 || 30.5 | ||
| Line 140: | Line 140: | ||
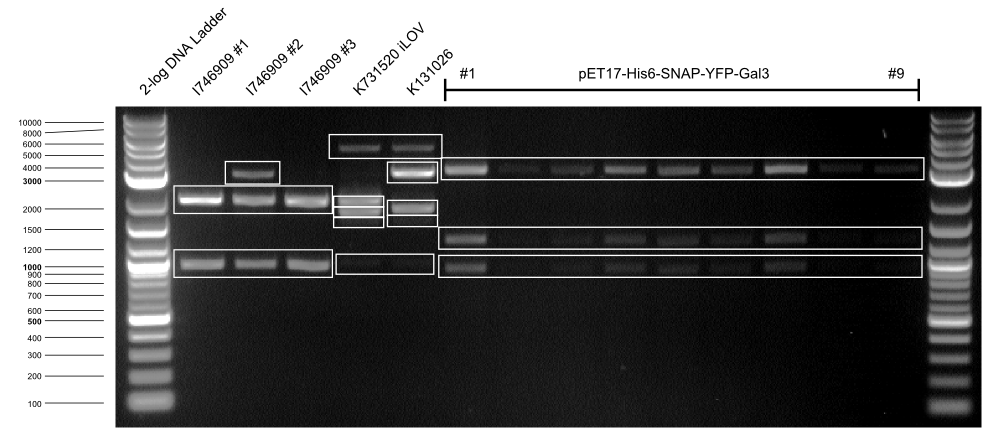
| I746909 BL21 #1 || 2029, 947 | | I746909 BL21 #1 || 2029, 947 | ||
|- | |- | ||
| - | | K1319042 | + | | K1319042 DH5α || 2029, 1780 |
|- | |- | ||
| - | | K131026 | + | | K131026 DH5α || 2029, 1848 |
|- | |- | ||
| pET17-Gal3 #1 || 3086, 923, 1262 | | pET17-Gal3 #1 || 3086, 923, 1262 | ||
| Line 158: | Line 158: | ||
* made alliquots of HM, 1 L HM + glucose + supplements and 500 ml LB | * made alliquots of HM, 1 L HM + glucose + supplements and 500 ml LB | ||
| - | * The | + | * The '''Quikchange''' mutagenesis PCR with a tamplate K1319000 and following primers was made. The PCR was made in two steps. The first step is PCR with both primers separatly and the second step includes PCR with two PCR products mixed with each other. |
** forward primer EYFPtoREACh1_F: AGTACAACTGGAACAGCCACAACGTCTATATC | ** forward primer EYFPtoREACh1_F: AGTACAACTGGAACAGCCACAACGTCTATATC | ||
** rewerse primer EYFPtoREACh1_R: GTTGTGGCTGTTCCAGTTGTACTCCAGCTTG | ** rewerse primer EYFPtoREACh1_R: GTTGTGGCTGTTCCAGTTGTACTCCAGCTTG | ||
| Line 203: | Line 203: | ||
<center> | <center> | ||
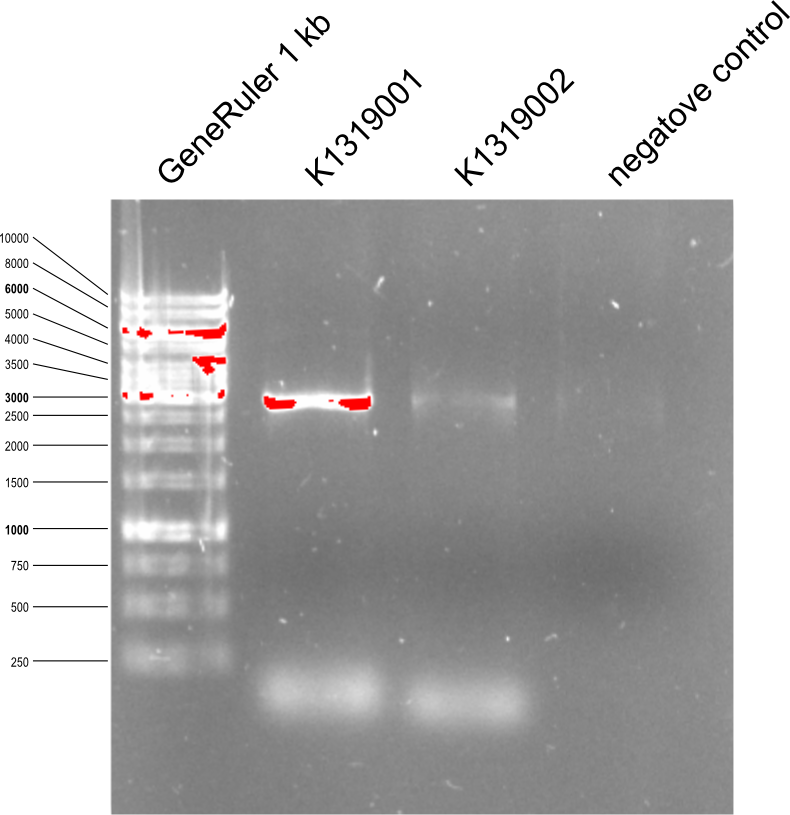
| - | {{Team:Aachen/Figure|Aachen_14-07-5_PCR_toREACh1%262.png|title=Agarose gel with PCR products after | + | {{Team:Aachen/Figure|Aachen_14-07-5_PCR_toREACh1%262.png|title=Agarose gel with PCR products after Quikchange from K1319000 to K1319001 and K1319002 |width=400px}} |
</center> | </center> | ||
* REACh1 gets number K1319001 and REACh2 gets number K1319002. | * REACh1 gets number K1319001 and REACh2 gets number K1319002. | ||
| - | * PCR product was restricted with DpnI | + | * PCR product was restricted with DpnI 60 min at 37°C to destroy the methylated template. Then DpnI was deaktevated at 80°C during 20 min. |
* PCR product was purifired and transformated in DH5α. | * PCR product was purifired and transformated in DH5α. | ||
== 6th == | == 6th == | ||
* transformation of J04450 in pSB1K3 and pSB1A3 in NEB10β cells. | * transformation of J04450 in pSB1K3 and pSB1A3 in NEB10β cells. | ||
| - | * | + | * plasmid prep of J04450 in pSB1C3, pSEVA234_LasR and pSEVA641_BsFbFP. |
| - | * made precultures of | + | * made precultures of NEB10β and DH5α cells |
* inoculation of the fermenter at 11:40, and induced the fermentation of pET17-Gal3. The fermentation is expected to run 24 h. | * inoculation of the fermenter at 11:40, and induced the fermentation of pET17-Gal3. The fermentation is expected to run 24 h. | ||
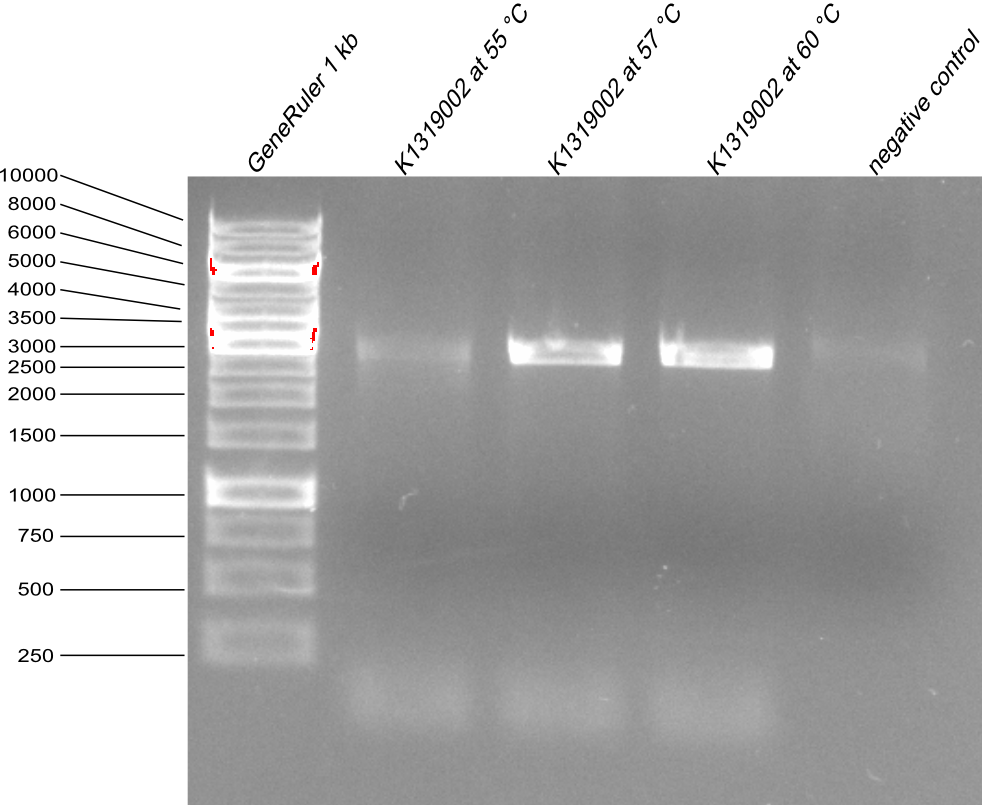
| - | * as the first Quickchange PCR for REACh2 was not sucsessful, it was reapeted as a gradient PCR with annealing temperatire | + | * as the first Quickchange PCR for REACh2 was not sucsessful, it was reapeted as a gradient PCR with annealing temperatire 55°C, 57°C and 60°C. Then the agarose gel with PCR product samples was made. |
<center> | <center> | ||
| Line 266: | Line 266: | ||
== 19th == | == 19th == | ||
* Chips with K131026 and I746909 in HM were made. Images were taken every 30 min with the Geldoc | * Chips with K131026 and I746909 in HM were made. Images were taken every 30 min with the Geldoc | ||
| - | * A PCR of J23101.E0240, K1319000, K1319001, K1319002 was run and the product was separated on a 1,2 | + | * A PCR of J23101.E0240, K1319000, K1319001, K1319002 was run and the product was separated on a 1,2% agarose gel |
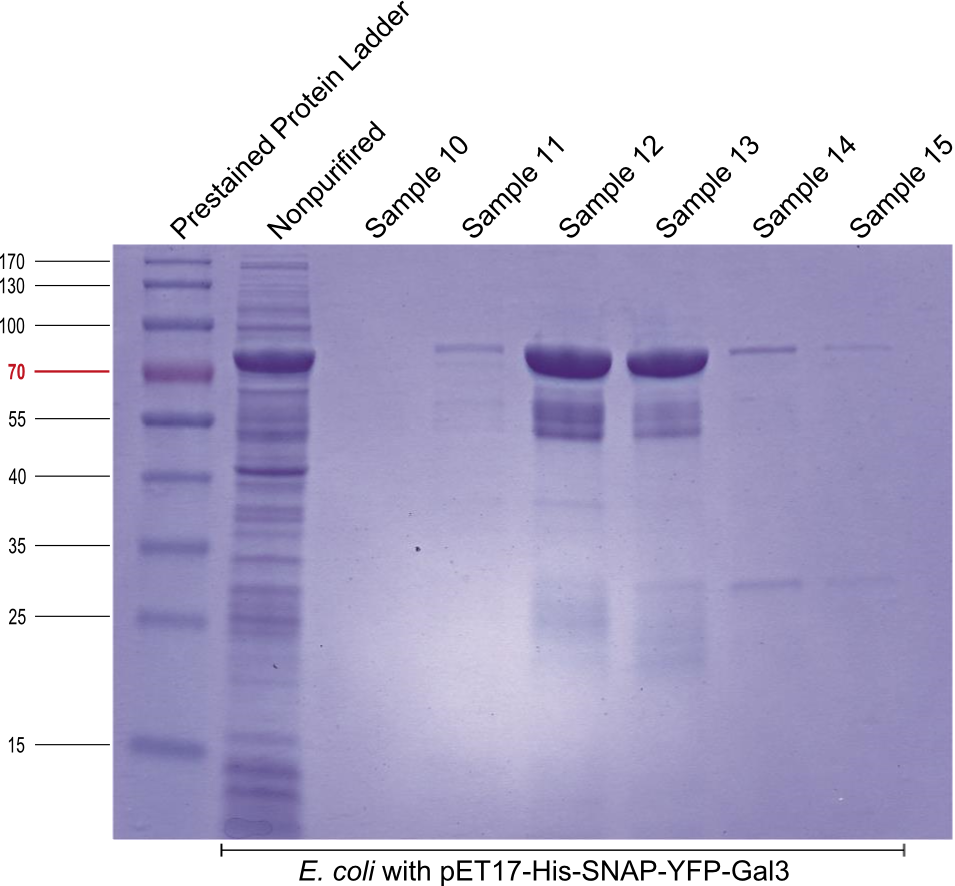
* A SDS gel with samples of protein purification of pET17-His-SNAP-YFP-Gal3 was made to check if the purifired samples contain the target protein - YFP-Gal3. The purification or the gel resulted in two samples with a yellow color. For the SDS we also used the two samples before and the two samples after ones with the yellow color, 6 mL. | * A SDS gel with samples of protein purification of pET17-His-SNAP-YFP-Gal3 was made to check if the purifired samples contain the target protein - YFP-Gal3. The purification or the gel resulted in two samples with a yellow color. For the SDS we also used the two samples before and the two samples after ones with the yellow color, 6 mL. | ||
| Line 284: | Line 284: | ||
* made chips of K131026 in NEB , DH5α and BL21 in LB and additionally K131026 in DH5α in HM+. Images were taken every 30 min with our own device | * made chips of K131026 in NEB , DH5α and BL21 in LB and additionally K131026 in DH5α in HM+. Images were taken every 30 min with our own device | ||
<center> | <center> | ||
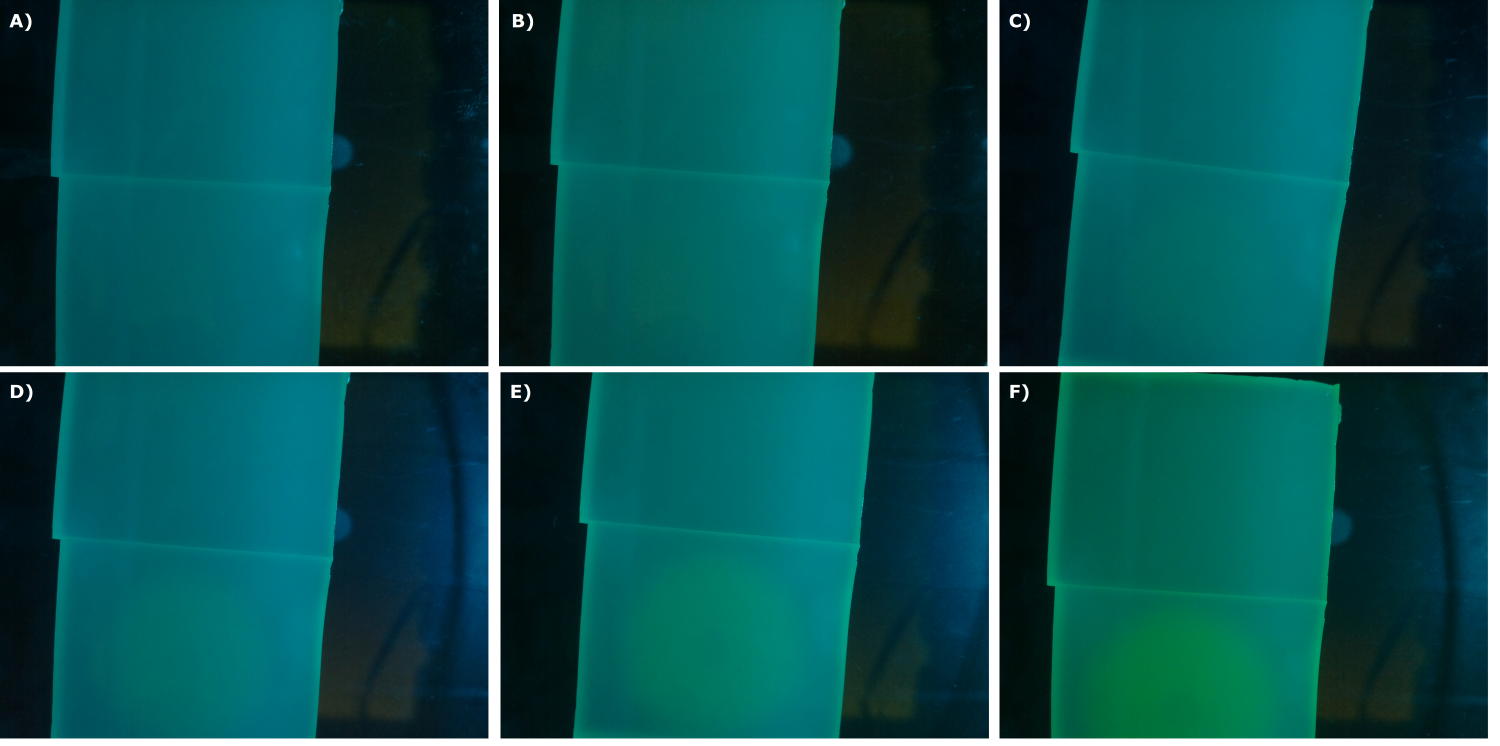
| - | {{Team:Aachen/Figure|Aachen_24_08_2014_K131026_bl21_serie.png|title=Sensor Chips with K131026 in BL21 in LB taken with first prototyp of our own device|subtitle=Sensor chips with K131026 in BL21 in LB medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_24_08_2014_K131026_bl21_serie.png|title=Sensor Chips with K131026 in BL21 in LB taken with first prototyp of our own device|subtitle=Sensor chips with K131026 in BL21 in LB medium with 1,5% agar, lower chip induced. A) befor induction with 2 µl of 5000 µg/ml HSL (3-oxo-C12) B) 0.5 h after induction C) 1 h after induction D) 1.5 h after induction E) 2 h after induction F) 2.5 h after induction|width=900px}} |
</center> | </center> | ||
== 25th == | == 25th == | ||
* did a plasmid restriction of I20260 (EcoRI,PstI), J23115 (EcoRI, SpeI), K516032 (XbaI,PstI), and J23101 (EcoRI, SpeI) | * did a plasmid restriction of I20260 (EcoRI,PstI), J23115 (EcoRI, SpeI), K516032 (XbaI,PstI), and J23101 (EcoRI, SpeI) | ||
| - | * tested the growth of ''Pseudomonas fluorescens'' in different liquid media for high OD and strong fluorescence. She tested Standard I medium, Cetrimide medium and Pseudomonas-F medium, and Pseudomonas-F medium supplemented with 300 µL Fe3+ in 500 mL flasks with a filling volume of 30 mL. The flasks were inoculated with ''P. fluorescens'' cells on Standard I agar, and incubated at | + | * tested the growth of ''Pseudomonas fluorescens'' in different liquid media for high OD and strong fluorescence. She tested Standard I medium, Cetrimide medium and Pseudomonas-F medium, and Pseudomonas-F medium supplemented with 300 µL Fe3+ in 500 mL flasks with a filling volume of 30 mL. The flasks were inoculated with ''P. fluorescens'' cells on Standard I agar, and incubated at 30°C at 250 rpm. |
* prepared over night cultures of K131026 in DH5α and NEB for chips | * prepared over night cultures of K131026 in DH5α and NEB for chips | ||
* prepared 2x 5 ml of pSB1C3, psB3K3 and pSB1A2 for plasmid prep | * prepared 2x 5 ml of pSB1C3, psB3K3 and pSB1A2 for plasmid prep | ||
| Line 301: | Line 301: | ||
* restriciton of synthesized TEV protease with EcoRI and PstI | * restriciton of synthesized TEV protease with EcoRI and PstI | ||
* qualitatively tested the ''Pseudomonas fluorescens'' that had grown over night for OD and fluorescence. She determined that Pseudomonas-F medium is the most adequate for the cultivation of the strain we use, since both OD and fluorescence were best in the flask containing the respective medium. Growth in the Pseudomonas-F medium supplemented with 300 µg/L Fe3+ was weaker, however, fluorescence was also successfully suppressed. | * qualitatively tested the ''Pseudomonas fluorescens'' that had grown over night for OD and fluorescence. She determined that Pseudomonas-F medium is the most adequate for the cultivation of the strain we use, since both OD and fluorescence were best in the flask containing the respective medium. Growth in the Pseudomonas-F medium supplemented with 300 µg/L Fe3+ was weaker, however, fluorescence was also successfully suppressed. | ||
| - | * made chips with K131026 in DH5α and NEB, in LB and LB + 10 | + | * made chips with K131026 in DH5α and NEB, in LB and LB + 10% glycerol. Images were taken with our own device every 10 min (illumination problems). |
<center> | <center> | ||
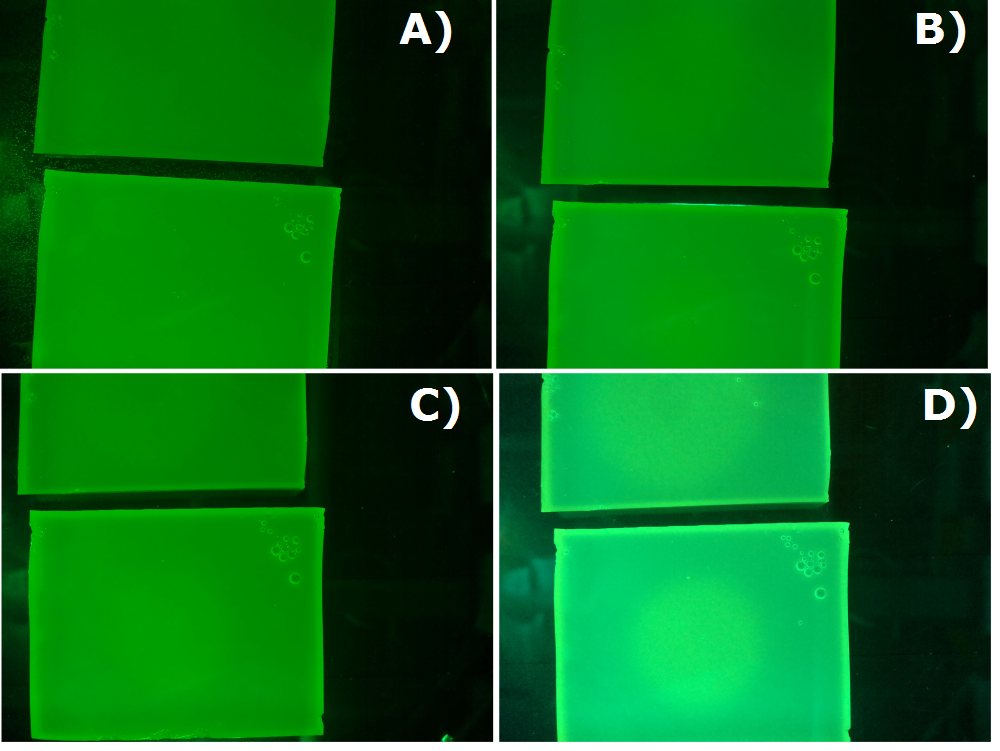
| - | {{Team:Aachen/Figure|Aachen_26_08_2014_K131026_neb_serie.png|title=Sensor Chips with K131026 in NEB in LB taken with first prototyp of our own device|subtitle=Sensor chips with K131026 in NEB in LB medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_26_08_2014_K131026_neb_serie.png|title=Sensor Chips with K131026 in NEB in LB taken with first prototyp of our own device|subtitle=Sensor chips with K131026 in NEB in LB medium with 1,5% agar. Chip on the top induced with 0.5 µl of 500 µg/ml HSL and on the bottom with less than 0.2 µl of 500 µg/ml HSL (3-oxo-C12). A) befor induction B) 1 h after induction C) 1.5 h after induction D) 2 h after induction |width=600px}} |
</center> | </center> | ||
* plasmid prep of the back bones, restriction and gel purification | * plasmid prep of the back bones, restriction and gel purification | ||
Latest revision as of 22:39, 17 October 2014
 "
"