Team:Heidelberg/pages/LOV/phototox
From 2014.igem.org
Contents |
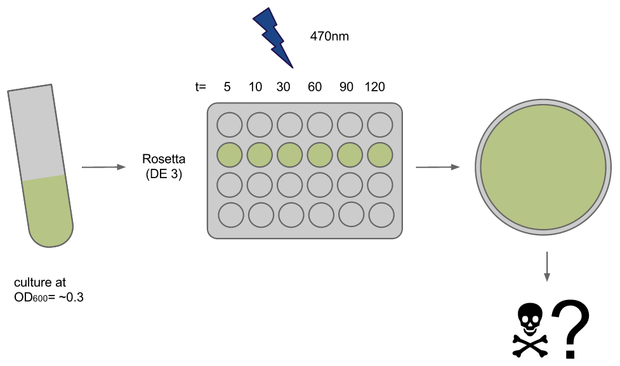
Expression strain phototoxicity
To determine if illumination with light of ~470nm had any toxic effect on our E. coli strains of choice we conducted a phototoxicity assay. Cultures were induced for different amounts of time and survival or rather their ability to grow was deduced from the number of colony forming units (CFUs) on agar plates after ~20h.
Dilution test
In a dilution assay bacteria were grown to OD600= ~0.8 and diluted 1:100, 1:10^4, 1:10^6 and 1:10^8. They were grown on LB + 1.5% agar plates at 37° overnight. Dilutions of 10^7 and 10^8 were chosen to be the most suitable for our readout as the resulted in 38 and 78 colonies with E. coli Rosetta and OneShot respectively.
Phototox 1
Cultures (500ul in 15ml each) of E. coli strains Rosetta and Oneshot were inocculated from o/n cultures and grown to OD= ~0.3
After transfer to a 24-well plate they were immediately induced in the Box at U=12V. Induction times were 5, 10, 30, 45, 60 and 120 min, the dark controls for each timepoint were kept tightly packed in aluminium foil.
For each timepoint the culture was transferred to an Eppendorf tube, diluted and plated onto LB+agar right after end of induction. Due to sudden shortage of time and agar plates some samples had to be kept on ice for ~1h.
Results
The experiment failed due to dilution errors and probably mixed up antibiotic resistances. Only the 10min timepoint showed any growth after 24h.
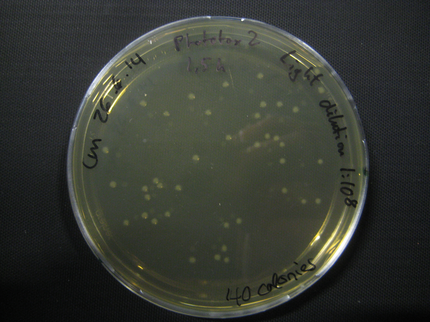
Phototox 2
Due to failure of the first assay the experiment was repeated with an easier setting using only the Rosetta strain and a dilution of 1:10^8.
The previous results (from Phototox 1) adumbrate that the Rosetta strain might already be affected by irradiation after 10 min. Therefore the Intensity was slightly decreased in the second experiment (U=11V instead of U=12V).
The assay was otherwise repeated in the same fashion as seen above. We were able to reduce the time from opening taking out the samples to having the bacteria plated to 6min.
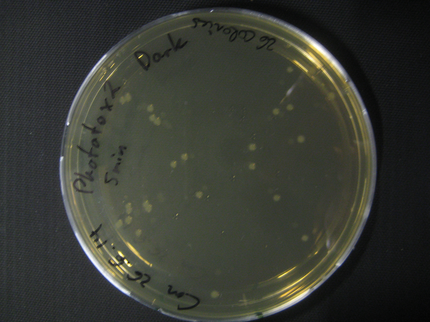
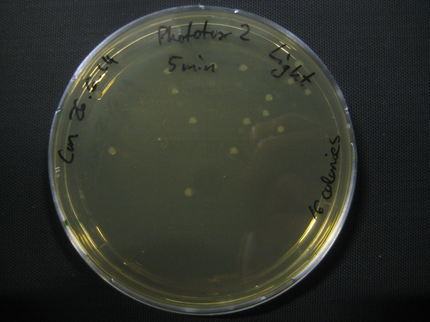
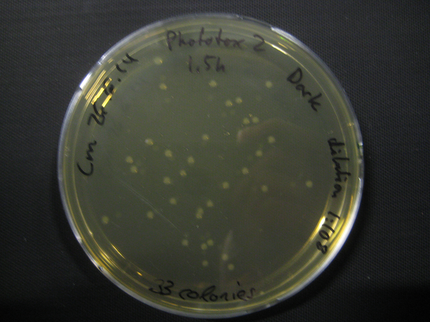
Results
| Time [min] | Light [CFUs] | Dark [CFUs] |
|---|---|---|
| 5 | 16 | 26 |
| 10 | 25 | 9 |
| 30 | 21 | 20 |
| 60 | 54 | 39 |
| 90 | 40 | 33 |
| 120 | full ( mistake in dilution) | full (mistake in dilution) |
Here we show pictures of two timepoints as examples.
LOV domain positive control
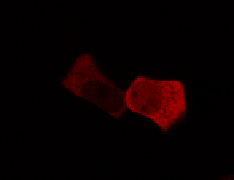
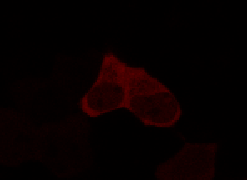
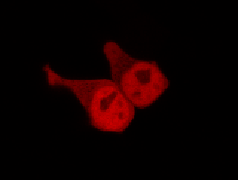
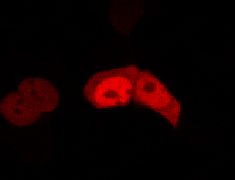
To determine if a conformational change in the As LOV2 domain we planned to use was actually triggered by our Induction Box we carried out an assay in a mammalian nuclear localisation system.
We used HEK 293 cells transfected with a construct containing a nuclear export signal (NES), mCherry, As LOV2 domain and a nuclear localisation signal (NLS) caged [1] in the J-alpha helix. The cells and the construct (Linus BiNLS 22) were obtained from Dominik Niopek [2].
Results
Confocal Microscopy clearly showed a light-dependent localisation to the cells nucleus.
References
[1] Strickland, D. et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 (2012).
[2] Niopek, D. et al. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat. Commun. 5, 4404 (2014).
 "
"