Team:SUSTC-Shenzhen/Project/gRNA-binding-target-sequence Plasmids Design and Construction
From 2014.igem.org
gRNA-binding-target-sequence Plasmids Design and Construction
Contents |
In this part, we have totally constructed 3 plasmids, to binding the gRNA sequences in human cells nucleus.
Abstract
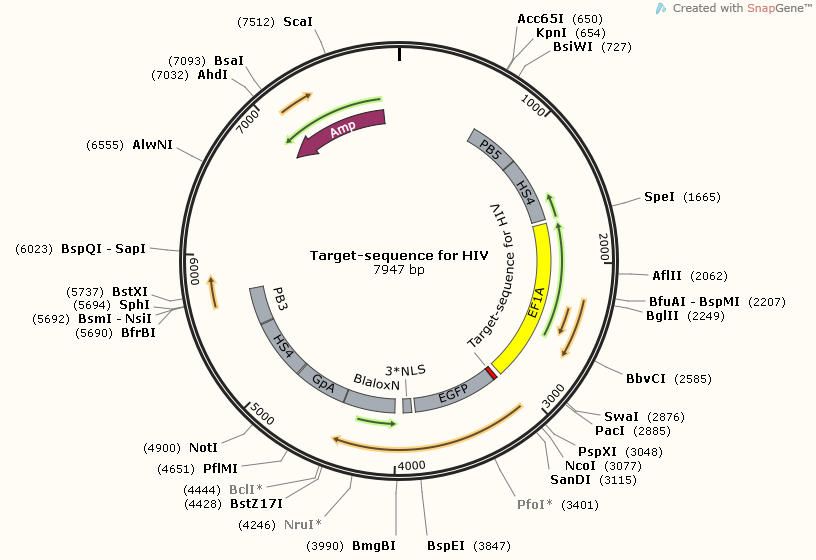
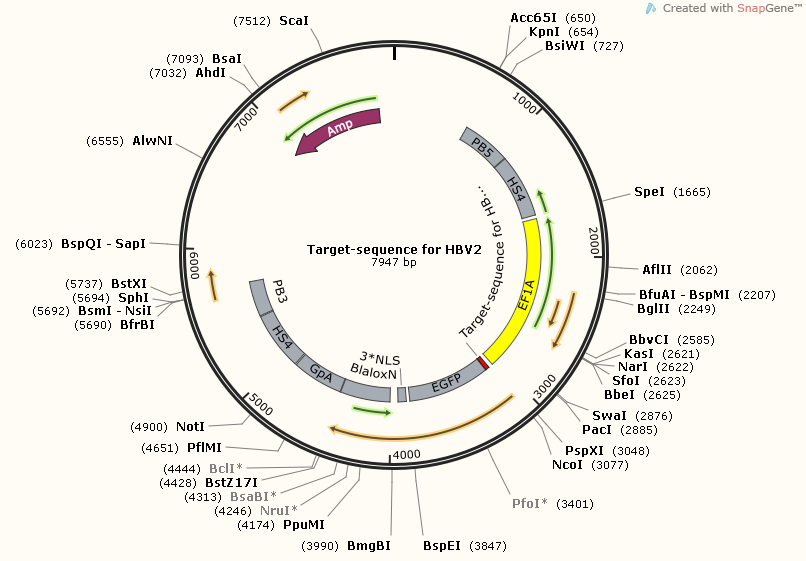
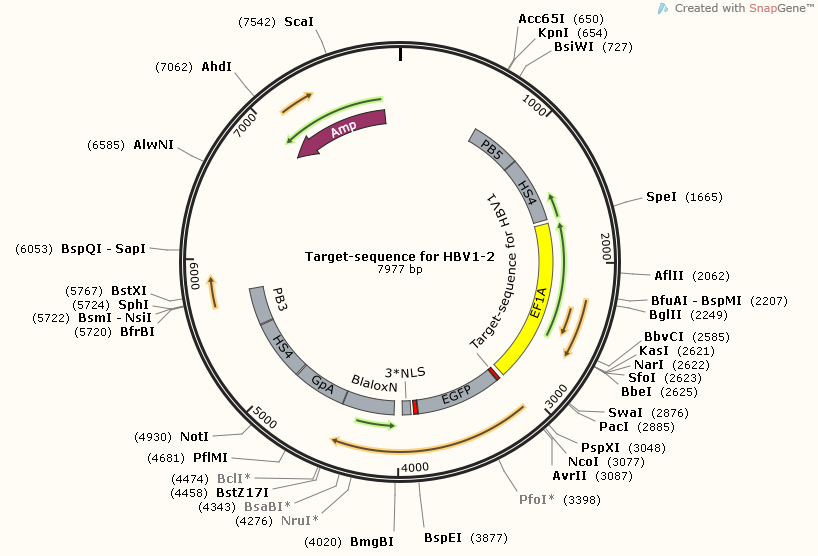
There are three target-sequence related plasmids. Each one corresponding a unique gRNA.
This three plasmid carry the Target-sequence aim to check if the CRISPR-Cas9 system we create in human cell can work well, which might be the foundation for the further experiments. .
However, although each one with different target-sequence, they still have plenty of components in common. You can see, all of them carry at least one section of target-sequences, and target-sequence for HBV1-HBV2 have two sections of target-sequence.This is because this part plays an important role in efficiency test, both for target-sequence binding gRNA and cut-off ability of CRISPR-Cas9.
Following are these three plasmids:
Part I Plasmid construction Design: Why do we construct them?
Why do we design plasmids that put target-sequence between promoter and EGFP?
As we all know,the double strands break of target-sequence is hard to observe in the human cell,it have no function for the normal activity of the cell,so we use the alternative solution.We insert the gRNA binding sequence between the promoter and reporter, transform the expression of DSB of binding sequence to the change of reporter. The whole target-sequence have a gRNA binding sequence in the middle and have more original sequence in two sides to return the natural state of HIV genome.
By this way, we can observe the DSB happen or not by the expression of EGFP.
Why dose each of our plasmids only carries sections of genome?
For safety,we can't use whole HIV or HBV genome in the lab, and actually the double strands break of genome is not vivid for quantitative.
In the alternative ,the fluorescence will directly link to the efficiency of double strands break,that's what we need.
Part II Method: How do we construct them?
Preparation of plasmid backbone:
All plasmids we constructed are based on the plasmid pBX-083PB5-HS4-EF1A-EGFPnuc-2A-Bla-LoxN-BGpA-HS4-PB3 (7896bp, HongKong) We use NcoI, BspEI to do enzyme digestion of plasmid pBX-083PB5-HS4-EF1A-EGFPnuc-2A-Bla-LoxN-BGpA-HS4-PB3, removing the entire EGFP sequence and collect the remaining 7179bp DNA fragment as ligation vector for the following experiments.
PCR for target-sequence from plasmid pBX-083PB5-HS4-EF1A-EGFPnuc-2A-Bla-LoxN-BGpA-HS4-PB3 (7896, HongKong):
First,we acquire our target-sequence locate between the promoter and EGFP by add target-sequence on primer.Second,we must mutate extra NcoI and BspEI enzyme site on the PCR product.
With primers A, we achieve:
Get the target-sequence with EGFP, also with NcoII and BspEI enzyme sites added on each end.
Primers target-sequence for HIV:
Forward (5’-3’): CATGCCATGGGATCAAAATCTCTAGCAGTGGCGCCCGAACAGGGACccttggtgagcaagggcgagg
Reverse (5’-3’): TACCTCCGGAGGGtccggtcttgtacagctcgtc
Primers target-sequence for HBV2:
Forward (5’-3’): CATGCCATGGGATCTCTCCTCTGCCGATCCATACTGCGGAACTCCTccttggtgagcaagggcgagg
Reverse (5’-3’): TACCTCCGGAGGGtccggtcttgtacagctcgtc
Primers target-sequence for HBV1-2:
Forward (5’-3’): CATGCCATGGGATCCCTAGGACCCCTGCTCGTGTTACAGGCGGccttggtgagcaagggcgagg
Reverse (5’-3’): CATGCCATGGGATCCCTAGGACCCCTGCTCGTGTTACAGGCGGccttggtgagcaagggcgagg
Construction of gRNA carried plasmids by Restriction enzyme digestion & Ligation:
Do enzyme digestion (with NcoI & BspEI) of PCR product.
Ligation for backbone and target-sequence. Thus, we get the target-sequence with EGFP plasmid.
Thus, we get the target-sequence carried plasmid:
TargetsequenceforHIV1-pBX-083PB5-HS4-EF1A-EGFPnuc-2A-Bla-LoxN-BGpA-HS4-PB3
TargetsequenceforHBV2-pBX-083PB5-HS4-EF1A-EGFPnuc-2A-Bla-LoxN-BGpA-HS4-PB3
TargetsequenceforHBV1-2-pBX-083PB5-HS4-EF1A-EGFPnuc-2A-Bla-LoxN-BGpA-HS4-PB3
Remark
The results of efficiency test for target-sequence binding gRNA transportation and CRISPR-Cas9 cut-off ability can be found in A-B toxin Part & Cell Culture Part.
 "
"