Team:Oxford/notebook
From 2014.igem.org
Lab Book
Part A
Part B
Week 1
Tasks completed:
Swapping resistance gene for plasmid pME6010 from tetracycline to kanamycin.
Methods:
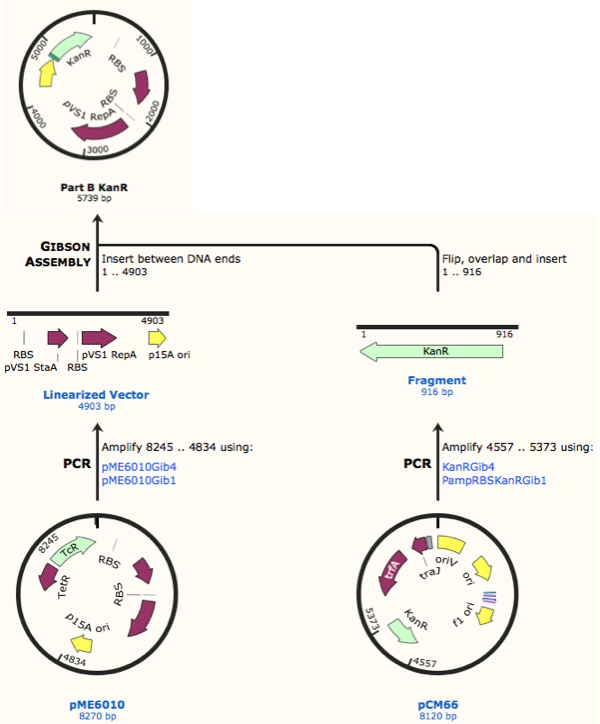
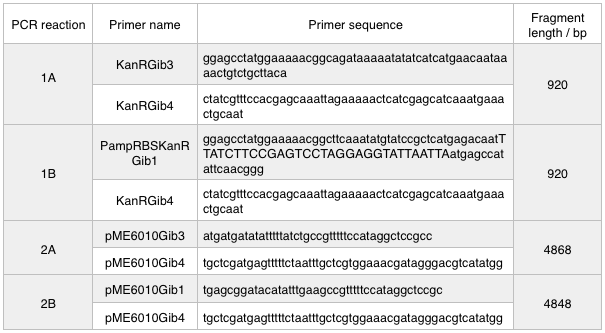
First, we designed forward and reverse primers for KanR (with native RSB and promoter) which we isolated from plasmid pCM66. The 5’ ends were complementary to the insert region of pME6010 (reaction 1).
We then designed forward and reverse primers to amplify the pME6010 backbone (reaction 2).
Further to this we redesigned each set of primers to incorporate an ampicillin promoter and optimised RBS (https://salis.psu.edu/software/) (B reactions) instead of the native promoter region (A reactions). This process is described in the following map:
The designed plasmids were as follows:
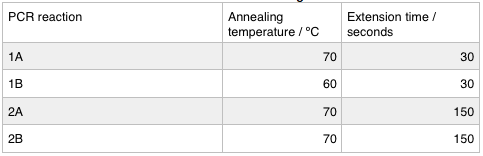
We ran these 4 PCR reactions as follows using the: NEB Q5 PCR protocol
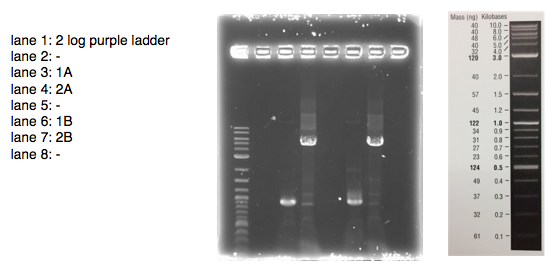
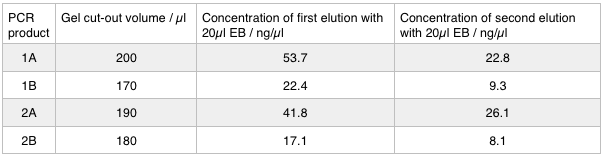
A 0.8% agarose gel was used for the extraction as this offered good separation around 1kb and 5kb. This gel was run and the bands extracted according to our QIAquick Gel Extraction Protocol using NEB purple loading dye and 2-log purple ladder and QIAquick extraction kit. Gel obtained:
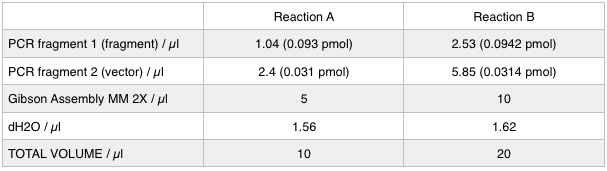
As we later found out these NanoDrop readings are false since the QG buffer in the QIAGEN gel extraction kit interferes with the UV/Vis readings. Following extraction of our PCR products from the gel we used the NEB Gibson Assembly protocol to run an 8hr reaction over night. We ran an ‘A’ reaction which will insert the KanR gene into the pME6010 plasmid with the native promoter and a ‘B’ reaction that will insert the KanR gene with a pamp promoter and optimised RBS.
The volumes were chosen to satisfy 100ng vector with a 3-fold increase in the amount of insert. The insert amount must lie between 0.02 and 0.5 pmol and the total volume of total fragments cannot exceed 10µl. Following an overnight 8hr Gibson Assembly the reaction volumes were treated with Dpn1 restriction enzyme that cuts bacterial (methylated) DNA. We transformed the Gibson products into chemically competent DH5-alpha cells as well as into NEB alpha-5 cells. Unfortunately no colonies grew on a KanR plate! We know that the PCR products are correct so we think an issue may have arisen during the Gibson Assembly stage so we will re-do this part next week.
 "
"