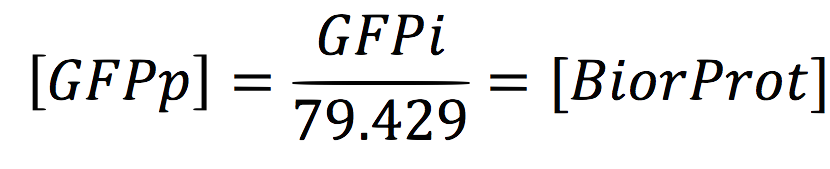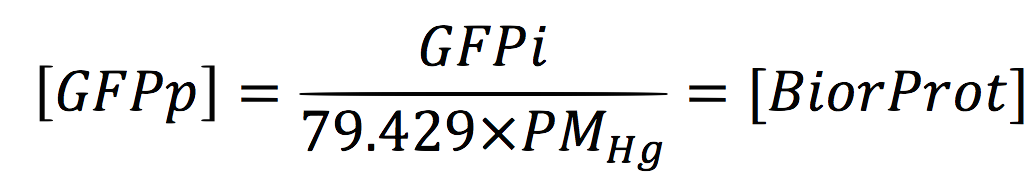Team:BIOSINT Mexico/collaboration
Collaboration
Promoter characterization
In order to treat atherosclerosis, the ITESM-CEM has developed the use of microbial enzymes that are able to degrade 7-ketocholesterol.5. We helped them characterize one of their promoters pCMV on yeast.
With Team Linköping University: Peanut allergies</p>
This team´s objective is to analyze food and drink to detect the Ara h allergen, helping peanut allergic consumers to make healthier and easier meal decisions. Their survey was held to examine how mexican society handles the needs of peanut-allergic people.
With ETH Zurich Team: Complexity patterns in nature</p>
We distributed and participated on this team´s survey to help them have broader demographics of the general public´s thoughts about complexity: either on handling the intricacy of many abstract concepts all together or finding this mystification in nature. All this involves a different perspective on how we find ourselves expressing and understanding complex patterns through our daylife. And relates to the project of this iGEM team by finding areas of opportunity where people are able to see patterns of their interest.
Wtih VirginiaTeam: Synthetic Biology Public Acceptance</p>
We interviewed 22 pedestrians in Queretaro to help the efforts of this team to understand how SynBio is regarded by the general public. Most of the questions were generally about GMOs, how the interviewed people would feel about consuming them and the implications that they would find the most compromising for safety.
With Team France: Weekly Newsletter</p>
The effort of this team is to recompile the updates of every iGEM team, making collaboration easier. Thus we send a brief description of our project to this team to collaborate on this weekly effort.
Predictive model for protein expression
As minery and urbanization byproducts concentrate and pollute some of our most precious resources, such as water, some of our peer iGEMers have designed solutions to remediate these hazards. Such is the case of UFAM_Brazil, whose project involves the integration of mercury resistance operon mer into a bacterial chassis. We were glad to involve ourselves with their mathematical modeling development...curious about the outcome? Here we show how it was resolved:
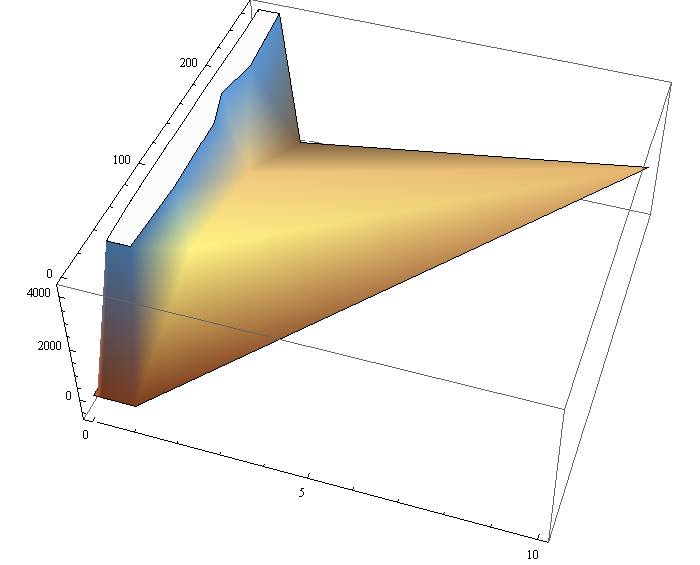
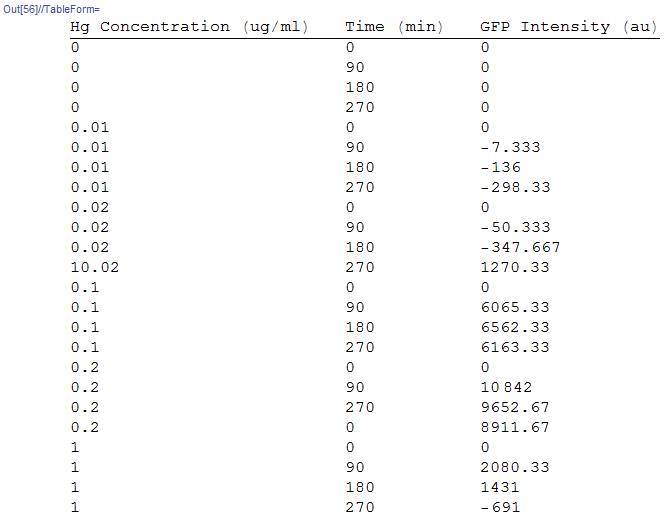
We use the GFP data for the factors concentration and time. Data was provided for specific experiments and we plotted the graphs for time vs concentration.
From this plot we infer that the equation that fitted our model would be and exponential equation. We transferred the data to the Mathematical software and organized the factors as time=x1 and concentration=x2.
Using a non-linear regression for the observe patterns. First we plot a 3d graph for the analyzed data and then we fed a model of the form.
Where GFPi is the data that we obtained, IC is the initial mercury concentration and t is the time.
Therefore, our equation for the fitted model is
With this equation we can infer the intensity of GFP given any time or any initial concentration of mercury, then using the equation reported by JCBRAFF(BBa_E0040) which is
Where y = GFP intensity and x = GFP concentration in nanomolar, we can infer the amount of GFP protein produced by the cell and finally have the ability to predict the concentration of our bioremediation proteins since the concentration of GFP is the same that this proteins. By the equation:
Since the GFPi is given in µg/ml, we have to divide the equation for the Mol weight of Hg.
 "
"