SubmittingFam20COct
From 2014.igem.org
Revision as of 20:32, 17 October 2014 by MariaChavez (Talk | contribs)
October 14, 2014: DNA pick-up by [http://sequetech.com/ Sequetech] to sequence 2 samples, detailed below
- Location: BioCurious
- Participants: Rachel (set up sample pick-up), Emi (gave samples to Sequetech rep)
- Notes: Rachel
- Aims: The Fam20C Kex + & Kex - inserts are too long to obtain complete reads with primers that anneal externally to the insert (see Submitting 10 casein and FAM20C in pD1214 plasmids for sequencing 30Sep2014.) To complete sequencing coverage, we are resubmitting these two constructs for sequencing with newly designed primers that anneal within the insert region.
- Sequencing Strategy & Methods:
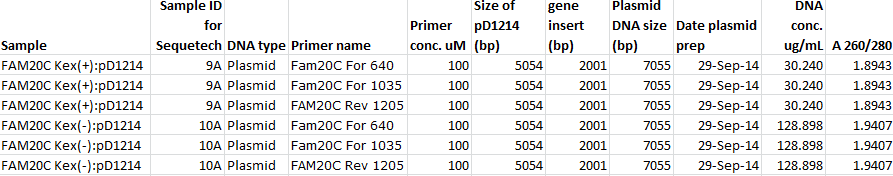
- Details about samples submitted for sequencing in table, below.
- Submitting 10uL aliquots of 2 samples: Sap.FAKS.hFam20C(Kex+).Sap:pD1214 and Sap.Faks.hFam20C(Kex-).Sap:pD1214 from Cloning bovine beta casein, Fam20C (Kex + & -), from 27Sep2014. Submitting clones 9A and 10A. Should have been 9B; 9A is "bad" per CR.
- Sequetech rep. collected samples at ~2:00pm today.
- All to be sequenced in three reactions: two forward primers (Fam20C For 640 & Fam20C For 1035) and one reverse (FAM20C Rev 1205). All primers designed 09Oct2014 by RVC project members.
Primer Sequences Fam20C For 640: 5' - CCTGCTTAGAGATCCA - 3' Binds internally in Sap.Faks.hFam20C(Kex + & Kex-).Sap gBlock DNA and reads through in the 5'-3' direction. Fam20C For 1035: 5' - ATTGATTATGACCTTCCA -3' Binds internally in Sap.Faks.hFam20C(Kex + & Kex-).Sap gBlock DNA downstream of Fam20C For 640 and reads through in the 5'-3' direction. FAM20C Rev 1205 : 5'-ATTCTACCAGCAACAG -3' Binds internally in Sap.Faks.hFam20C(Kex + & Kex-).Sap gBlock DNA and reads through in the 3'-5' direction.
Details for 6 sequencing reactions on plasmid DNA from 2 clones:
October 15, 2014
Sample 10A sequenced perfectly. Read to be transformed into yeast!
- Need to submit FAM20 Kex + clone 9B for this full-coverage sequencing.
 "
"