Team:SUSTC-Shenzhen/Project
From 2014.igem.org
Project Description
What is our story?
Contents |
Abstract
In this project, we want to establish a more effective HIV-curing system with less side-effects by integrating CRISPR/Cas system into human hematopoietic stem cells, which aims to protect the helper T cells from virus infection. The gRNA is designed to target the relative conserved regions in HIV viral genome and inactivate its biological activity. Since viral vectors seem to be of limited use in gene therapy strategies (e.g., potential pathogenicity), there is still a need for a simple, efficient nucleic acid transfer system which allows the target-cell-specific introduction of nucleic acids. By using non-viral DNA delivery system like A-B-toxin-GAL4 fusion protein, we can deliver plasmids encoding gRNA into the helper T cells and readily attack multiple HIV genome sites simultaneously and update the targets along with our knowledge of HIV.
Background
HIV (Human immunodeficiency virus)
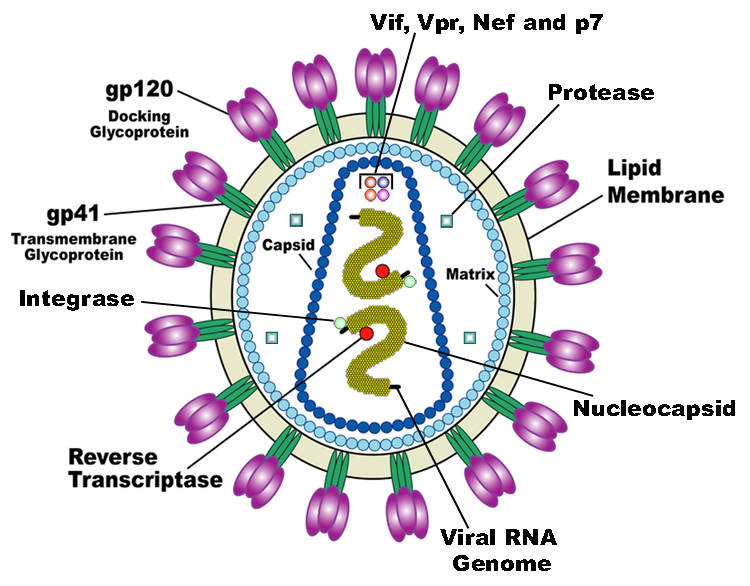
HIV is the trigger of the lethal disease AIDS. It replicates in and kills helper T cells, weakening human immune system and allowing life-threatening opportunistic infection and cancers to thrive. HIV infects vital cells in the human immune system such as helper T cells (especially CD4+ T cells), macrophages, and dendritic cell. As a retrovirus, HIV reverse transcribes its genome and integrated it into the host cell genome after infection. Figure 1 demonstrates the schematic structure of HIV virus (Source: Wikipedia).
Traditional therapies like chemotherapy or radiotherapy can’t protect the helper T cells from virus invasion and the virus can lurk in the helper T cells; drug treatments like HAART (Highly Active Antiretroviral Therapy) can only control the development of the disease and they also have strong side-effects. The process of reverse transcription is extremely error-prone, and the resulting mutations may cause drug resistance or allow the virus to evade the body's immune system.
CRISPR/Cas system
CRISPR/Cas (Clustered regularly interspaced short palindromic repeats/CRISPR-associated system) is originally a self-protecting mechanism in bacteria against external DNA such as virus genome or plasmids. It uses CAS complex to cut the external viral or plasmid DNA and integrating a short DNA segment into the CRISPR loci in the bacteria genome. This short DNA segment will be transcribed into pre-crRNA and bind to another CAS endonuclease called Cas9 with the help of tracrRNA. The Cas9 endonuclease cuts the external DNA in the existence of not only crRNA but also a special recognizing sequence at the 3’ end of the target DNA called PAM (Protospacer Adjacent Motif). When the same external DNA invades next time, the Cas9-tracrRNA-crRNA complex will recognize and cut it in order to destroy its biological activity. Figure 2 demonstrates the mechanism of Type II CRISPR/Cas system in bacteria (Mali, Esvelt, & Church, 2013). Dr. George Church (Mali, Yang, et al., 2013) and Dr. Feng Zhang (Cong et al., 2013) both have successfully transfected Type II CRISPR-Cas system into human cells, which is the foundation of this project.
Figure 2 Mechanism of Type II CRISPR/Cas system in bacteria
c) A-B toxin
A-B toxin are two-component exotoxin secreted by a number of pathogenic bacteria. The complexes contain two subunits called A subunit and B subunit. A subunit is the active portion that is poisonous to host cells, and is transferred to the host cell through endosomes mediated by the B subunit. The B subunit is a translocation and binding protein. Figure 3 shows how Anthrax toxin enter into human cells.
Figure 3 Model of Anthrax toxin entering into human cells d) PiggyBac (PB) transposon system PiggyBac (PB) transposon is a mobile genetic element that efficiently transposes between vectors and chromosomes via a "cut and paste" mechanism but leave no “footprint”. PBase in this system recognizes the transposon-specific inverted terminal repeat sequence (PB5 & PB3 in Figure 6) located on both end of transposon vector and efficiently moves the contents from the original sites and efficiently integrates them into TTAA chromosomal sites. Figure 4 demonstrates the working mechanism of PB transposon system (Source: Wikipedia).
Figure 4 Mechanism of PiggyBac transposon system e) Tet-ON 3G operon system The Tet-On 3G Systems are inducible gene expression systems for mammalian cells. Target cells that express the Tet-On 3G transactivator protein and contain a gene of interest (GOI) under the control of a TRE3G promoter (PTRE3G) will express high levels of your GOI, but only when cultured in the presence of doxycycline (Dox), which is a synthetic tetracycline derivative. The TRE3G promoter is very sensitive to the concentration of doxycycline and functions like a step function after induction. Also, the Dox concentrations required for induction of Tet-On Systems are far below cytotoxic levels for either cell culture or transgenic studies. So it is ideal to use Tet-ON 3G as a mammalian gene expression controller. Figure 5 demonstrates the mechanism of Tet-ON 3G system.
Figure 5 Mechanism of Tet-ON 3G operon
Content
You can use these subtopics to further explain your project:
- Overall project summary
- Project Details
- Materials and Methods
- The Experiments
- Results
- Data analysis
- Conclusions
It's important for teams to describe all the creativity that goes into an iGEM project, along with all the great ideas your team will come up with over the course of your work.
It's also important to clearly describe your achievements so that judges will know what you tried to do and where you succeeded. Please write your project page such that what you achieved is easy to distinguish from what you attempted.
 "
"