Team:NCTU Formosa/results
From 2014.igem.org
Our experiment can roughly divided into two categories.
1. About E.coli. Aspects:gene recombination and protein expression.
2. About Insect Aspects:PBAN effect testing、insects' habbit testing and our device testing
Contents |
E.coli. Aspects
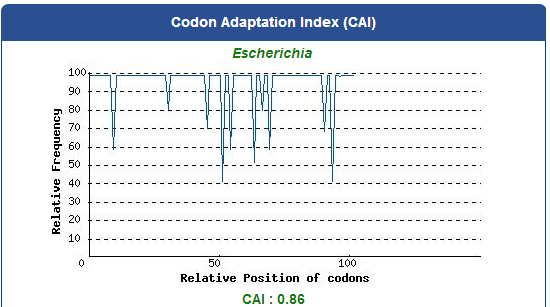
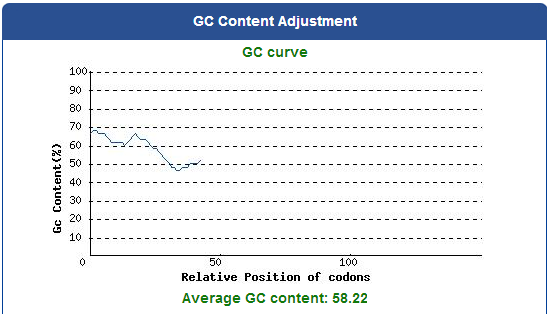
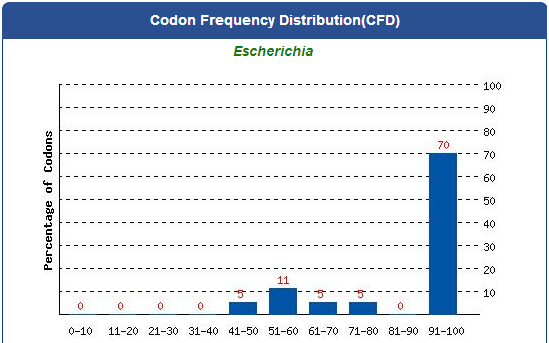
DNA Synthsis of 9 different kinds of PBAN
For our project (aim for capturing harmful insects) this year, we first found 9 different kinds of PBAN peptide of common agricultural harmful insects in the world from many reference papers. Then, we used these peptides to surf the NCBI and found the DNA sequence from the certain insect. ( EX:PBAN Spodoptera litura:http://www.ncbi.nlm.nih.gov/protein/AAK84160.1 ) Finally, we modified every codon on the DNA sequence and designed the DNA sequence for our E.coli. to express a certain PBAN.
DNA Modification Process:
1. Avoid the rare codon of E.coli., and choosing high frequency codons.
( Frequence Table Tool:http://www.genscript.com/cgi-bin/tools/codon_freq_table )
2. Don't choose the same codon to modify our designed gene many times, or our E.coli. will not have enough nucleotides to replicate it.
3. Avoid the start codon ATG existing in the front of our DNA sequence.
4. Use Rare Codon Analysis Tool ( http://www.genscript.com/cgi-bin/tools/rare_codon_analysis ) to inspect if there is any problem to express our gene
for E.coli.
Take PBAN Spodoptera litura for example:
5. Add iGEM standard sequence in front of and at the back of our modified DNA sequence.
6. Let the gene synthesis company synthesize our modified DNA sequence.
PCR for 9 different kinds of PBAN

For checking the size of the DNA sequence getted from the gene synthesis company, we recombined each PBAN gene to PSB1C3 backbone and do PCR ro check each PBAN size.
Because our PBAN DNA sequence length is around 100~150 bp., the PCR result should be 415~515 bp. The fig.2-1-3 shows the correct size of our PBAN, and proves we succeed in ligating our PBAN gene to the ideal backbone.
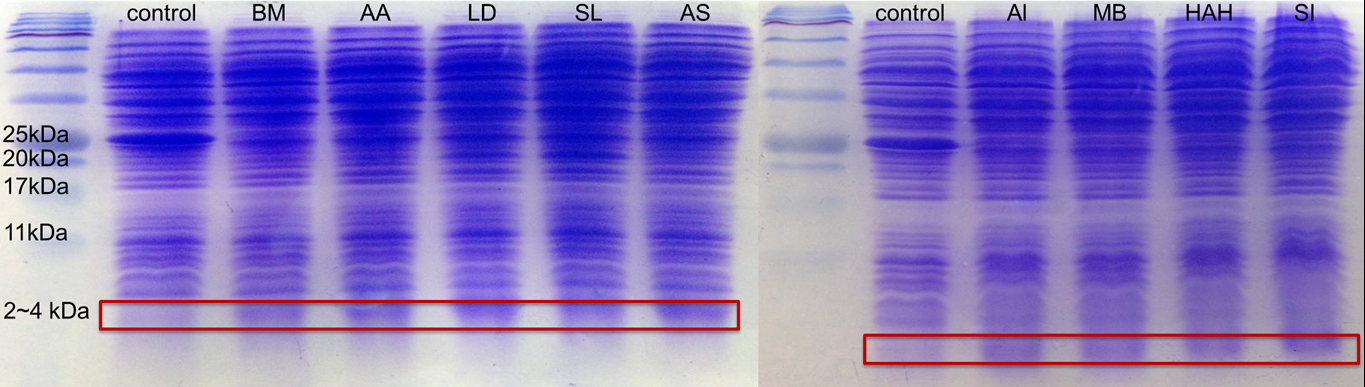
SDS Protein Electrophoresis of 9 different kinds of PBAN

In advance, for proving all the 9 different kinds of PBAN can be produced by our E.coli., we smashed the E.coli.,containg our PBAN with Sonicator and take the supernatant divided from the bacterial pellet after centrifuged to do 20% gel SDS protein electrophoresis.
Because PBAN peptide is an around 30 amino acids substance, in fig.2-1-5 we can see the band at 2~4kDa, and the E.coli. containg Pcons+RBS Plasmid don't. This result proves that our E.coli. can really produce our PBAN.
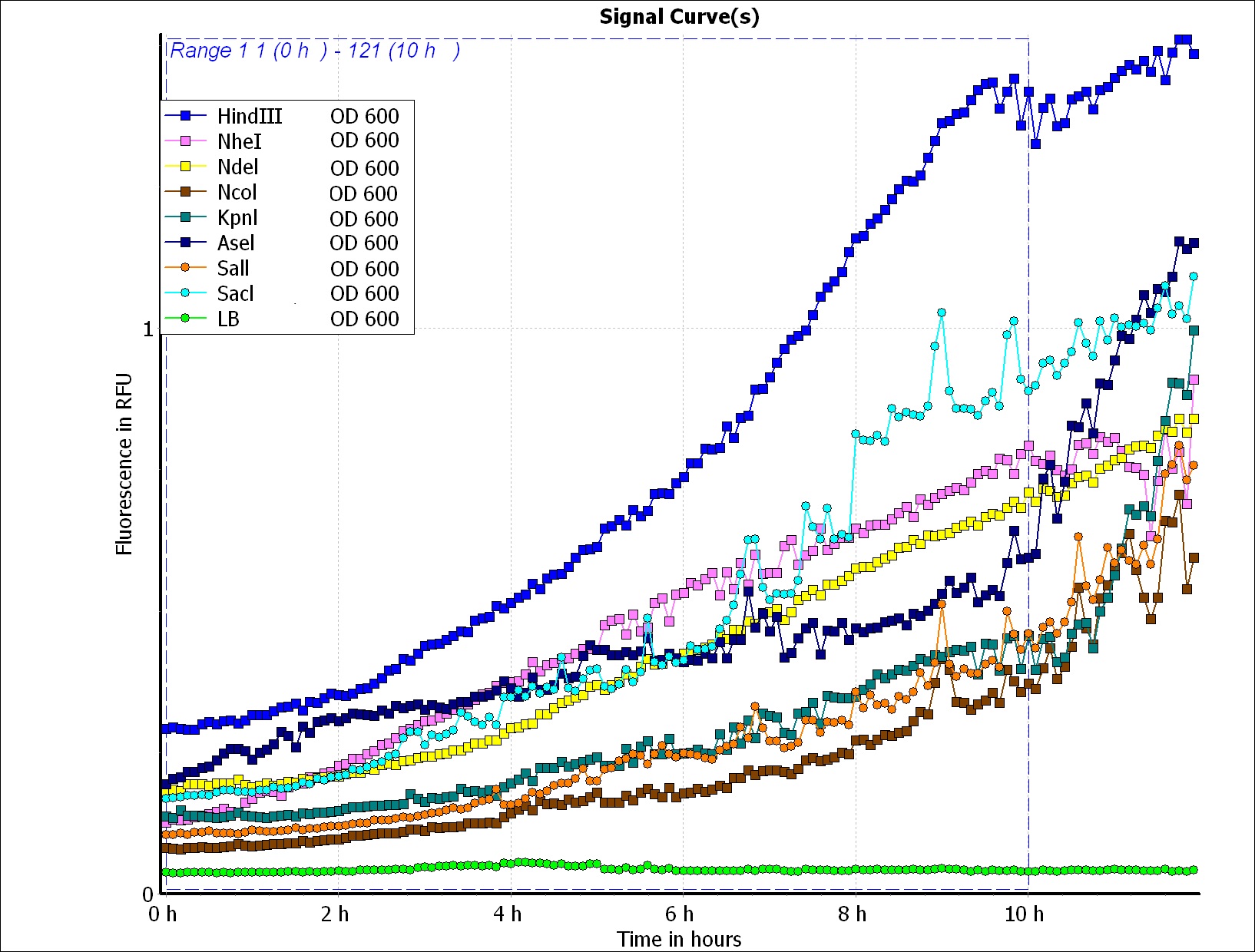
BFP Fluorescence / OD 600 Bacteria Growth Testing

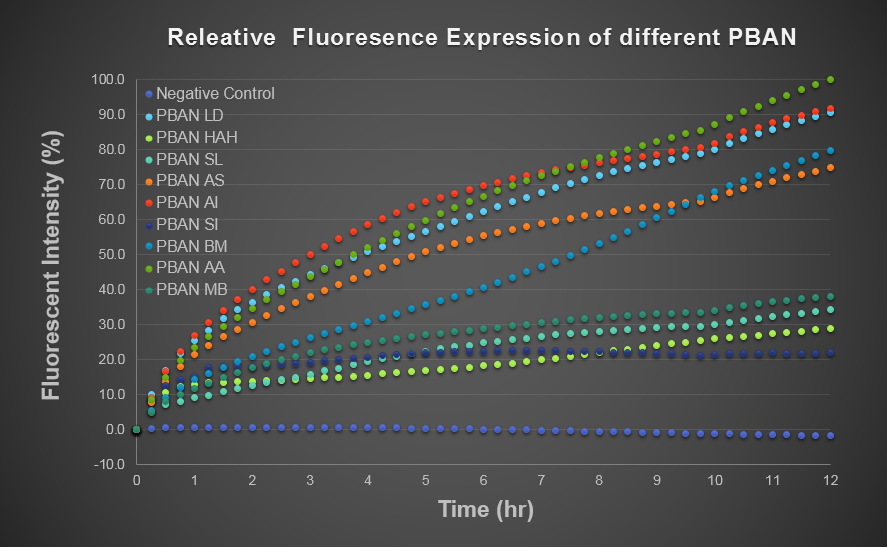
We wanted to use computer modeling to help us predicting PBAN expression in E.coli., so we decided to use BFP flourescence to reach our goal. Our thought is that we can compute the average value of the BFP flourescence expression value of the biobrick part (above) and of the Pcons + RBS + BFP + Ter and regard the average value as the prediction of our PBAN expression in E.coli. ( Pcons + RBS + PBAN + Ter ) </div></div>( More detail can be seen in our Modeling Page. ) This is the BFP flourescence expression curve and bacterial growth curve (OD 600) below in long time ,we use these data to predict our PBAN expression in E.coli..
Insect Aspects
Behavior of Target Insects Eating Our PBAN
Effect Testing of Our PBAN
Testing of Spodoptera Litura Hobby for Temperature and Light
Magic Power of Our Pyramidal Device
 "
"