Team:Paris Saclay/Notebook/July/21
From 2014.igem.org
(Difference between revisions)
(→2 - Results: Transformation of DY330 via pJBEI6409) |
(→Monday 21st July) |
||
| Line 4: | Line 4: | ||
===1 - Results: Transformation of supercompetent cells with CaCl<sub>2</sub>=== | ===1 - Results: Transformation of supercompetent cells with CaCl<sub>2</sub>=== | ||
| - | |||
''by Romain'' | ''by Romain'' | ||
| Line 13: | Line 12: | ||
===2 - Results: Transformation of DY330 via pJBEI6409=== | ===2 - Results: Transformation of DY330 via pJBEI6409=== | ||
| - | |||
''by Sean'' | ''by Sean'' | ||
| Line 21: | Line 19: | ||
|+Number of colonies per dish | |+Number of colonies per dish | ||
|- | |- | ||
| - | ! | + | ! 50μl from cuvette |
| - | ! | + | ! 100μl |
! remainder | ! remainder | ||
|- | |- | ||
| Line 28: | Line 26: | ||
| 4 | | 4 | ||
| 30 | | 30 | ||
| - | ! | + | ! 2μl of plasmid |
|- | |- | ||
| 3 | | 3 | ||
| 6 | | 6 | ||
| 45 | | 45 | ||
| - | ! | + | ! 4μl |
|- | |- | ||
| 0 | | 0 | ||
| Line 42: | Line 40: | ||
===3 - Oligo's design=== | ===3 - Oligo's design=== | ||
| - | |||
''by Romain'' | ''by Romain'' | ||
===4 - Liquid Bacterial Culture=== | ===4 - Liquid Bacterial Culture=== | ||
| - | |||
''by Marie, Romain & Sean'' | ''by Marie, Romain & Sean'' | ||
| Line 54: | Line 50: | ||
===5 - Electrophoresis=== | ===5 - Electrophoresis=== | ||
| - | |||
''by Fabio (process A) and Mathieu (process B and C)'' | ''by Fabio (process A) and Mathieu (process B and C)'' | ||
| Line 61: | Line 56: | ||
[[File:LU000070.jpg|right]] | [[File:LU000070.jpg|right]] | ||
| - | We used | + | We used 2µl of DNA of the following Biobricks in a 1% Agarose Gel. |
''The PCRs came from Mathias' manipulation made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/18 18th July].'' | ''The PCRs came from Mathias' manipulation made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/July/18 18th July].'' | ||
Revision as of 13:49, 22 July 2014
Contents |
Monday 21st July
Lab Work
1 - Results: Transformation of supercompetent cells with CaCl2
by Romain
Strains transformed: MG1655, MG1655Z1. from Bacterial Cultures transformed the 18th July
Results: Nothing has grown.
2 - Results: Transformation of DY330 via pJBEI6409
by Sean
Transformation performed on the 18th July
| 50μl from cuvette | 100μl | remainder | |
|---|---|---|---|
| 0 | 4 | 30 | 2μl of plasmid |
| 3 | 6 | 45 | 4μl |
| 0 | 0 | 0 | Control |
3 - Oligo's design
by Romain
4 - Liquid Bacterial Culture
by Marie, Romain & Sean
- DY330 pJBEI6409 with 10µl Cm in 10ml LB (x2)
- BT340 Cm and Amp
- The new strains received
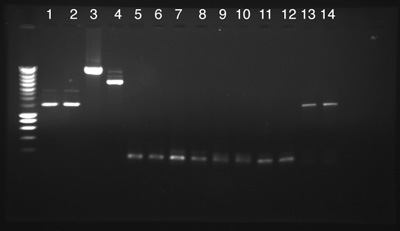
5 - Electrophoresis
by Fabio (process A) and Mathieu (process B and C)
We used 2µl of DNA of the following Biobricks in a 1% Agarose Gel. The PCRs came from Mathias' manipulation made the 18th July.
Process A
- J23119 Cl1
- J23119 Cl2
- J23106 Cl1
- J23100 Cl2
- PCR 1
- PCR 2
- PCR 3
- PCR 4
- PCR 5
- PCR 6
- PCR 7
- PCR 8
- PCR 9
- PCR 10
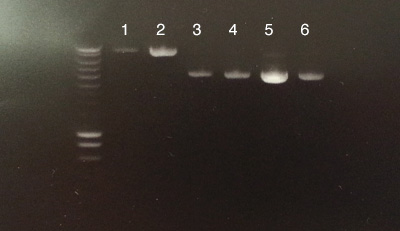
Process B
Process C
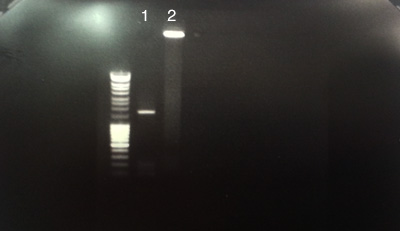
Pooling and purifying PCR 9 and 10 from process A.
- PCR purified products. IPS70 / IPS71 of pOsV230 (Apramycin)
- BT 340 (plasmid's flipase)
Results:
- A: From 1 to 4: Success, DNAs have the expected size.
- A: From 5 to 12: Failure, No PCR products.
- A: Numbers 13 and 14: Success, PCR products have the expected size.
- B: All 6 extractions were successful.
- C: Number 1: successful concentration of pOsV230's PCR product.
- C: Number 2 had no migration.
People there:
- Instructors and advisors: Solenne and Sylvie.
- Students: Arnaud, Fabio, Juliette, Mathieu, Marie, Pierre, Romain, Sean and Terry.
 "
"