Team:Imperial/coculture
From 2014.igem.org
| Line 105: | Line 105: | ||
<div class="pure-u-1-1"> | <div class="pure-u-1-1"> | ||
<h3>Results</h3> | <h3>Results</h3> | ||
| - | <figure class="content-image image- | + | <figure class="content-image image-full"> |
<img class="image-full" src="https://static.igem.org/mediawiki/2014/1/19/IC14-coculture-fig5.png"> | <img class="image-full" src="https://static.igem.org/mediawiki/2014/1/19/IC14-coculture-fig5.png"> | ||
<figcaption>Figure 5: the ratios shown for the graph legend indicate the ratio of glycerol to glucose in the carbon feedstock. a) Level of E. coli growth in HS media RFP co-culture with the <em>G. xylinus iGEM</em> strain for different feedstocks, b) Thickness of the pellicle in the HS media RFP co-culture indicating the growth of the <em>G. xylinus iGEM</em> strain with a dummy scale of 1-5 being used where 5 is a very thick cellulose pellicle. </figcaption> | <figcaption>Figure 5: the ratios shown for the graph legend indicate the ratio of glycerol to glucose in the carbon feedstock. a) Level of E. coli growth in HS media RFP co-culture with the <em>G. xylinus iGEM</em> strain for different feedstocks, b) Thickness of the pellicle in the HS media RFP co-culture indicating the growth of the <em>G. xylinus iGEM</em> strain with a dummy scale of 1-5 being used where 5 is a very thick cellulose pellicle. </figcaption> | ||
Revision as of 21:30, 17 October 2014
RFP E. coli and G. xylinus iGEM Co-Culture
Overview
Based on the hypothesis of E. coli BL21D3 operating anaerobically and G. xylinus replicating aerobically, a co-culture experiment for bacterial cellulose was attempted. Initially, the best possible carbon source among 5 available was found experimentally. This data was used to inform the choice of glycerol and glucose as the carbon feedstock. With these carbon feedstocks, E. coli with RFP expressed were successfully embedded in the bacterial cellulose, and RFP was clearly visible.
Key Achievements
- Experimentally determined the optimum carbon feedstocks for a HS media based co-culture of E. coli and G. xylinus.
Introduction

The combination of E. coli as an efficient cloning organism with a large library of well characterised parts and G. xylinus as a robust efficient cellulose producing host came about as a way to take advantage of the characteristics of each host. E. coli DH10B has a very short division time of 30 min whereas the G. xylinus iGEM strain has been producing bacterial cellulose robustly in a co-culture with yeast. For these reasons, the aim was to have E. coli produce customisable proteins of interest such as metal binding phytochelatin, fused to cellulose binding domains (CBD), simultaneously with cellulose being produced by G. xylinus allowing these proteins to attach to the material, effectively making it a 1 step functionalisation.
In a co-culture of the cellulose-producing and protein-producing species there will be competition for resources and space, and the potential of a symbiosis. The main hypothesis of this co-culture experiment is that G. xylinus would orient itself in the oxygen rich layers towards the top of the media due to being an obligate aerobe and E. coli would orient itself throughout the media but expectedly have a higher concentration in the oxygen depleted bottom media.
In order to have a stable co-culture, at least in the timescales we are conducting our growth and functionalisation, we need to identify potential interactions and regulate the growth of each species. The main variable identified for the interactions was identified as the carbon source of the HS media used. This requires an initial experiment that can inform the choice/combination of carbon source(s) used for the actual experiment containing E. coli with an Anderson promoter controlled RFP gene (J23104, make this hyperlink to http://parts.igem.org/Part:BBa_J23104) in J61002, (link this to http://parts.igem.org/Part:BBa_J61002) and the G. xylinus iGEM strain.
Preliminary Experiment
Methods
In terms of measurements, the doubling rate of E. coli of 30 min whilst the doubling rate of G. xylinus is approximately 8 hours, which means that by far the major contribution to total population, and so OD600, is E. coli. Therefore, the OD600 will be used as an indication of the E. coli level in the media. For G. xylinus, the cellulose pellicle is assumed to be a direct indication of the population of G. xylinus. Triplicates were used for all carbon feedstocks and HS media was prepared according to the G. xylinus HS media and culturing protocol. A 2 %w/v concentration of carbon source was used. The Kombucha media was prepared according to the Kombucha growth protocol.
15 ml Falcon tubes were inoculated with 12 ml of media, the samples were seeded with a small solid seed of pellicle of the G. xylinus iGEM strain. Lids were kept loosely unscrewed to allow aeration, and samples were incubated at 30°C static. Triplicates were used for all carbon feedstocks and a control of unseeded media was incubated alongside the samples.
Results

Shown in figure 2 are feedstocks that allowed growth of both E. coli and G. xylinusE. coli in the media. Figure 2 b) depicts reasonable in G. xylinus growth in glucose, acetic acid, glucose & glycerol 50/50 combination and glycerol. Taking this data into account, glucose & glycerol would be the best candidates for a co-culture. However, the optimum ratio of glucose to glycerol has not been discovered, so 6 combinations of glycerol to glucose ratios would have to be tested.
TThe samples depicted in figure 3 illustrate qualitatively the significance of carbon source for the co-culture. The 50/50 glucose glycerol based HS media displayed compartmentalisation which can be explained by specialisation of E. coli and G. xylinus according to oxygen levels as hypothesised.
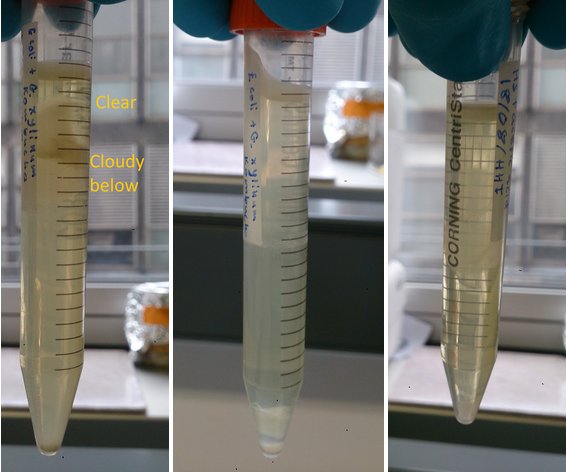
Anderson RFP E. coli and G. Xylinus iGEM co-culture
Methods
To set up the co-cultures, an RFP expressing colony was picked from a plate containing J23104 in J61002 and incubated in 300 ml of Ampicillin LB media in a 1000 ml conical flask overnight. Triplicates were set up for 6 different ratios (100/0, 80/20, 60/40, 40/60, 20/80 and 0/100) of glycerol to glucose with the sum of the two being equal to a total 5 %w/v of carbon source. 100 ml conical flasks were set up with 40 ml of HS media, 900 ul of E. coli and a large physical seed of G. xylinus iGEM was added. The seeds were taken from the initial co-culture experiment’s pellicles formed in glucose, glycerol and glucose & glycerol feedstock experiment. The samples were incubated at 30°C, static.

Results



Some conical flasks slowly developed expression of RFP in the bacterial cellulose. The ratios of glycerol to glucose in the HS media that first showed expression of RFP on the pellicle was 80/20 as illustrated in figure 6.
After a longer incubation period, the expression of RFP increased as the cell population increased as shown in figure 7.
Discussion
The results indicate that embedding E. coli into bacterial cellulose is most efficient with HS media containing 2 %w/v glycerol and 3 %w/v glucose. This showed promising intercalation of E. coli in the cellulose and could therefore provide a means to create 1 step functionalised cellulose.
Limitations occur in terms of potential contamination, and with respect to control of population. Control of population may be achieved by adjusting the level of acidity of the medium since the pH-growth curves for G. xylinus and E. coli may be different (variation of pH tolerance between different strains). Both can survive in a range of pH from low (~3) to moderate (~8), but the exposure time may differ. E. coli briefly faces the low pH of the stomach and duodenum, while G. xylinus has a longer period of acidic conditions as it secretes gluconate and acetate. Therefore, it may be possible to regulate the faster-growing E. coli simply using the pH of the medium rather than additional factors such as feedstock.
Moreover, the carbon source preference of each species could also be a point of population control, e.g. if one uses a specific source much less easily or efficiently than the other, or not at all. For example, G. xylinus strains.
 "
"











