Team:NTNU Trondheim/Project/Modelling
From 2014.igem.org
| Line 255: | Line 255: | ||
<p> | <p> | ||
To aid in the identification of possible target genes for increasing CO<sub>2</sub>uptake in Synechocystis, <a href="http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1003081">the most recent model of the metabolic network in Synechocystis</a> was acquired and analyzed with | To aid in the identification of possible target genes for increasing CO<sub>2</sub>uptake in Synechocystis, <a href="http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1003081">the most recent model of the metabolic network in Synechocystis</a> was acquired and analyzed with | ||
| - | <a href="http://www.nature.com/nbt/journal/v28/n3/abs/nbt.1614.html">flux balance analysis</a>. The approach initially chosen was simply to fix the CO<sub>2</sub> uptake flux above that achieved in the optimal FBA solution, and examining the effect on the growth rate (figure 1). | + | <a href="http://www.nature.com/nbt/journal/v28/n3/abs/nbt.1614.html">flux balance analysis</a>. Computation was performed it the <a href="http://opencobra.sourceforge.net/openCOBRA/Welcome.html">COBRA toolbox</a> in Matlab 2014a.The approach initially chosen was simply to fix the CO<sub>2</sub> uptake flux above that achieved in the optimal FBA solution, and examining the effect on the growth rate (figure 1). |
<div class="col4"><a href="https://static.igem.org/mediawiki/2014/0/0b/Growth.jpg"> </a><img src="https://static.igem.org/mediawiki/2014/0/0b/Growth.jpg" width="500"> | <div class="col4"><a href="https://static.igem.org/mediawiki/2014/0/0b/Growth.jpg"> </a><img src="https://static.igem.org/mediawiki/2014/0/0b/Growth.jpg" width="500"> | ||
Revision as of 21:20, 17 October 2014
Team:NTNU_Trondheim/Home
From 2014.igem.org
Modelling
Flux balance analysis of the impact of increasing CO2-uptake and introducing Glucose Oxidase into Synechocystis sp. PCC6803
To aid in the identification of possible target genes for increasing CO2uptake in Synechocystis, the most recent model of the metabolic network in Synechocystis was acquired and analyzed with flux balance analysis. Computation was performed it the COBRA toolbox in Matlab 2014a.The approach initially chosen was simply to fix the CO2 uptake flux above that achieved in the optimal FBA solution, and examining the effect on the growth rate (figure 1).
The growth rate increases from 0 as the CO2 uptake flux is increased from zero to its optimal value (1.19), and decreases after this point. The decrease in growth rate after this optimal CO2 uptake flux is linear with a slope of -0.0083. As the growth rate decreases with increasing CO2 fixaton after this point, this means that any metabolic reaction added to the model that increases the CO2 fixation without also increasing the growth rate, will reduce the growth rate by at least -0.0089/h per unit of flux the CO2 uptake is increased by.
The glucose oxidase gene was added to the model in order to examine its effect on cellular growth. This was done by implementing the reaction:

Since the metabolite D-glucono-1,5-lactone was not present in the original metabolic model, this issue was addressed by the addition of a passive export reaction. The flux through this reaction was then increased from 0, and the CO2 uptake predicted by the FBA solution recorded (figure 2)
The FBA solution then predicts that the CO2 uptake increases as the glucose oxidase flux increases. In accordance with the previous observation that the reduction in growth rate would be at least -0.0089/h per per unit of the flux the CO2 uptake was increased by, the reduction in growth rate was found to be -0.0533 per unit of CO2 uptake flux.
Simulation of genetic circuit
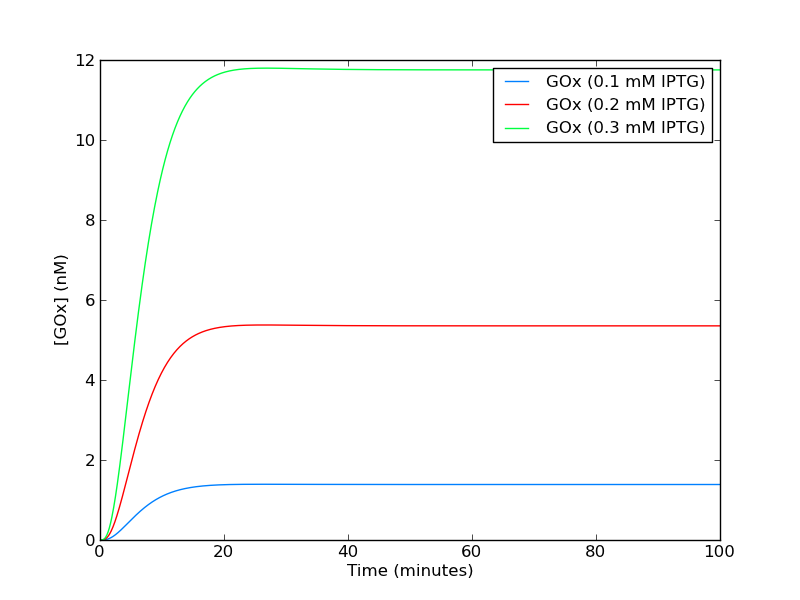
The construct chosen to express glucose odxidase (GOx) is a lac promotor regulated by LacI. GOx is therefore inducible by the addition of IPTG. The genetic circuit can be modelled by the following chemical reaction equations:

The system was simulated deterministically using Cain with the parameters in table 1. Parameters were taken from [1].
Table 1: Parameters for the inducible circuit
| Reaction rate constant | Propensity | Comment |
|---|---|---|
| k1 | 0.23 nM minute -1 | Transcription of lacI mRNA |
| k2 | 15 minute -1 | Translation of LacI |
| k3 | 0.5 | Transcription of GOx mRNA |
| k4 | 30 minute -1 | Translation of GOx |
| k5 | 0.92 minute -1 | Passive IPTG influx |
| k-5 | 0.92 minute -1 | Passive IPTG efflux |
| k6 | 50 nM -1 minute -1 | LacI dimerization |
| k-6 | 1e-3 minute -1 | LacI dimer dissociation |
| k7 | 3e-7 nM-2 minute -1 | LacI dimer IPTG binding |
| k-7 | 12 minute -1 | LacI dimer IPTG unbinding |
| k8 | 960 nM-1 minute -1 | Repression of GOx |
| k-8 | 2.4 minute -1 | Derepression of GOx |
| d1 | 0.2 minute -1 | Degradation of GOx |
| d2 | 0.2 minute -1 | Degradation of LacI |
| d3 | 0.462 minute -1 | Degradation of GOx mRNA |
| d4 | 0.462 minute -1 | Degradation of LacI mRNA |
| d5 | 0.2 minute -1 | Degradation of LacI dimer |
| d6 | 0.2 minute -1 | Degradation of LacI dimer-IPTG complex |
A .zip file with the model file can be found here. Simulating the system with various levels of IPTG (figure 3) yields increasing steady state levels of GOx.
 "
"
 "
"