Team:BGU Israel/Notebook1
From 2014.igem.org
| Line 107: | Line 107: | ||
<h3 style="border-bottom:dashed;border-color:#000000">May</h3> | <h3 style="border-bottom:dashed;border-color:#000000">May</h3> | ||
<br> | <br> | ||
| - | <div align="center" style="float:left" class="prevNext" onClick="show( | + | <div align="center" style="float:left" class="prevNext" onClick="show(april)"><img src="https://static.igem.org/mediawiki/2014/8/8f/BGU14finger-point3.gif" width="26px"/> Prev</div> |
| - | <div align="center" style="float:right" class="prevNext" onClick="show( | + | <div align="center" style="float:right" class="prevNext" onClick="show(june)">Next <img src="https://static.igem.org/mediawiki/2014/d/dd/BGU14finger-point.gif" width="26px"/></div> |
<div class="clear"></div> | <div class="clear"></div> | ||
<div class="textContNotebook" style="height:460px; background-image:background-image:"https://static.igem.org/mediawiki/2014/2/21/BGU14notebook.png""> | <div class="textContNotebook" style="height:460px; background-image:background-image:"https://static.igem.org/mediawiki/2014/2/21/BGU14notebook.png""> | ||
| Line 785: | Line 785: | ||
</section> | </section> | ||
| - | |||
Latest revision as of 15:36, 17 October 2014


Click on the Month or Week to check out what we did 
| April | May | June | July | August | September | October |
|---|---|---|---|---|---|---|
| Week 2 | Week 6 | Week 10 | ||||
| Week 3 | Week 7 | Week 11 | ||||
| Week 4 | Week 8 | Week 12 | ||||
| Week 1 | Week 5 | Week 9 | Week 13 |
April
 Prev
Prev
Lab
Our Ideas:
- Exocytosis of proteins by E.Coli for treating diseases- especially Diabetes.
- Citrus greening- bacterial disease of plants
- Skin cancer
- Early detection of Salmonella- preventing food poisoning
- Modified yeast for longer shelf life of bread
- Malaria- one of the largest death causes
- Bacterial producing of insulin in the intestine
- Methicillin-resistant staphylococcus aureus (MRSA)
MALARIA it is!-Lets build a project!
Research is going on!- Reading, talking, arguing and asking
General
- Looking for the team
- Interviewing students for the team- Finalizing team roster after interviewing
- FIRST MEETING!-Getting to know each other, What is iGEM competition? Thinking about project ideas
- Meeting with Human Practice team leader of 2013 - Learned about the nature of Human Practice and what its stands for, and got a better understanding of their thinking process about Human Practice strategies.
Starting reading and looking for previous iGEM outstanding projects at the Human Practice field. Maybe
May
 Prev
Prev
Lab
- Are we in the right direction? A lot of dead ends came up and made as wonder if we made the right choice for our research subject.
- Malaria is OUT- Metabolic Syndrome- IN: After the disappointment, we came up with a new idea!
- Talking about our reading conclusions
- Genetic Engineering for non-biologist: Introduction of the world of synthetic biology to our non- biologist team members, with a simple words
- Research around the clock!- Reading a lot, looking after experts for consulting
Human Practice
Meeting with the chairman of the BGU Students Union – Mr. Avi Ben Hillel
21/5- Presenting our project concept to the BGU Board of Governors
Meeting with Jill Ben-Dor and David Spivak from the department of Donor and Associate Affairs for promoting our project.
Meeting with Yossi Shavit, Director of Bengis Center for entrepreneurship & Hi-Tech Management. Planned our panel event on the innovation day 2014.
We need a LOGO!
The 44 annual Board of governors meeting- Presenting our idea, vision and plan for the first time, in front of the board of governors
June
 Prev
Prev
Lab
- Getting to know the lab- First time at prof. Smadar Cohen's lab, getting familiar with the equipment, facilities and common method we intend to use in our research
- The strategies:
1. Silencing the mRNA of PTP1b protein
2. Adiponectin overexpression
3. PGC1-a modest expression circuit
Human Practice
Meeting with Assaf Rudich
18/6/14- Producing an expert panel on innovation day 2014– "The Metabolic Syndrome - Could Synthetic Biology Provide a Breakthrough Solution"?
Meeting with Deans of Our faculties- Dean of Natural Sciences Faculty, Prof. Jiwchar Ganor; Dean of Engineering Sciences Faculty, Prof. Joseph Kost; and Dean of Humanities and Social Sciences, Prof. David Newman.
Meeting with Dr. Ariel B. Lindner from Paris Descartes University – Understood the meaning of Human Practice more deeply, shaped our HP strategies according to his notes.
Innovation day- "The Metabolic syndrome, Diabetes and obesity"
July
 Prev
Prev
Lab
- Liquid medium transfering
- mRuby DNA extraction- miniprep protocol
Human Practice
Starting working on our project video, meeting with the producer and came up with the brand "Inner Doctor".
Week 1: July 27th - August 2nd
 Prev
Prev
Lab
Intelligent Medication
Propagation of pcDNA3-mRuby2
Starters were taken from a pcDNA3-mRuby2 bacterial stab (acquired form Addgene) and incubated in LB containing 100 µg/ml ampicillin for 24 hours at 37?c. Plasmid DNA was extracted from using a miniprep kit.
Human Practice
Meeting with Dean of students, Prof. Moshe Kaspi and presenting our project.
August
 Prev
Prev
General
*****
Week 2: August 3rd- August 9th
 Prev
Prev
Lab
Intelligent Medication
Validation of co-expression of mRuby2 and eGFP in CT26 eGFP cell line
Goal: A preliminary experiment to validate we can visualize the co-expression of both mRuby2 and eGFP in CT26 eGFP cell line using confocal microscopy. Method: 20,000 cells were plated in each well of a chamber slide and incubated for 24 hours at 37?c. 4 wells were transfected with pcDNA3-mRuby2 using Lipofectamine 2000 (protocol), and incubated for 48 hours at 37?c. The other 4 wells were mock transfected with Lipofectamine 2000 and incubated in the same conditions for negative control. After 48 hours, the transfected cells were viewed at the confocal microscope. eGFP: excitation - 488 nm, emission - 509 nm mRuby2: excitation - 559 nm, emission - 600 nm
Results:

Co-expression of mRuby2 and eGFP. All three pictures show the same cells. Picture A shows only eGFP expressing cells, picture B shows only mRuby2 expressing cells, and picture C show all cells.
Conclusions:
mRuby2 and eGFP were co-expressed in CT26 eGFP cells and viewed successfully using confocal microscopy.
Human Practice
Meeting with the CEO of the Diabetes Israeli Association, Mr. Moti Perlmutter; discussed our project and learned important facts and statistic information about our target audience - diabetics (an acute part of the Metabolic Syndrome).
Shaping our Human Practice strategies according to the meeting and came up with a final approach.
Week 3
 Prev
Prev
Lab
No Activity
Human Practice
Meeting with Campaign Manager Mr. Guy Seemann- discussed our Bedouin Campaign
Week 4
 Prev
Prev
Lab
No Activity
Human Practice
No Activity
Week 5: August 31th-September 6th
 Prev
Prev
Lab
Intelligent Medication
Functional silencing experiment
Goal: Silencing eGFP in CT26 eGFP cells co - transfected with pcDNA3 mRuby2 and our scRNA construct. The scRNA construct is designed to be activated by mRuby2 mRNA, becoming a dicer substrate and finally silence eGFP. Method: We gave a different treatment for each of our four groups:

We conducted the experiment both in 24 well plate (for flow cytometry) and in a chamber slide (for confocal microscopy).
The cells were plated on the appropriate plates for the different tests (100,000 cells per well in the 24 well plate and 15,000 cells per well in the chamber slide) and incubated for 24 hours at 37?c.
After 24 hours, the incubated cells were transfected (according to the different treatments detailed above) with one or more of the following:
- pcDNA3 mRuby2 - 1.0 µg
- scRNA – both detection part A + hair pin part B - 100 pmol for each of the parts (A+B)
Transfection was done by lipofectamine (protocol), and then the cells were incubated for 48 hours at 37?c.
Preparations for image stream were done by the general protocol (protocol).
Results:
Image stream (flow cytometry):
Treatment 1 – Negative control

Treatment 2 – transfected with pcDNA3 mRuby2

Treatment 3 – transfected with scRNA

Treatment 4 – transfected with pcDNA3 mRuby2 & scRNA

Confocal microscope:
Co-expression of mRuby2 and eGFP was validated in all relevant wells.

Conclusions:
- GFP negative population: The first parameter to compare that comes to mind when looking for silencing is the percentage of GFP negative cells of the entire population – if a significantly bigger portion of the population is GFP negative, then the silencing worked. However, when looking at the population distribution of the different control groups, we can see no significant difference in the GFP negative population. This might not the result of unsuccessful silencing – we are using a stable GFP expressing cell line, but no matter how stable it is, always there will be a portion of the cells that do not actually express GFP. Additionally, a successful silencing is not expected to completely shut down the expression of GFP in single cells. This creates the necessity to evaluate the expression of GFP using another parameter.
- Mean intensity of GFP positive cells: This parameter will show difference between a negative control and a successful silencing in light of the above. However, cells treated with scRNA showed only a very small difference in this parameter, so we could not observe a successful silencing.
For the next try, the following changes were implemented:
- double the amount of scRNA in the transfection
- Change the protocol of preparing Lipofectamine – scRNA mix to allow for better interaction between the 2 parts of the construct, and prepare the Lipofectamine- DNA mix separately.
Human Practice
No Activity
September
 Prev
Prev
General
*****
Week 6: September 7th-September 13th
 Prev
Prev
Lab
Intelligent Medication
Goal: A second try of silencing eGFP in CT26 eGFP cells co - transfected with pcDNA3 mRuby2 and our scRNA construct. The scRNA construct is designed to be activated by mRuby2 mRNA, becoming a dicer substrate and finally silence eGFP.
Details:
The changes concluded from former results:
- Half of calls amount was seeded (50,000 cells per well)
- The cells were transfected with double amount of scRNA:
- pcDNA3-mRuby2- 1.0 µg
- scRNA – both detection part A + hair pin part B- 200 pmol for each of the parts (A+B)
- This time, the lipofectamin mixes were prepared separately for the pcDNA3-mRuby2 and for the scRNA (both A and B parts together), so that both of the mixes will make its content ready and prepared for transfection.
The rest of the protocol remained unchanged (protocol).
Notes:
During this transfection a mistake occurred. In one of the plates, an incorrect of volume of Lipofectamine-scRNA was added due to a measurement error. Consequently, this plate had unknown concentrations of nucleic acids and lipofectamine, with a positive deviation in volume.
Results:
Most of the cells died, probably due to the increased volume of Lipofectamine that was added by mistake.
Biobricks
Goal: The ordered parts for Biobricks preparation had arrived in the weekend so preparations of plates and medium for the Biobricks construction were done.
Details:
- LB agar plates with ampicillin (100 µg/ml) or Chloramphenicol (25 µg/ml) were made following the LB agar plates protocol.
- SOC medium for bacterial transformation was prepared. SOC medium content is:
- 0.5% (w/v) yeast extract
- 2% (w/v) tryptone
- 10 mM NaCl
- 2.5 mM KCl
- 20 mM MgSO4
- Glucose 20%
Preparation followed by the SOC medium prep protocol.
Human Practice
Meeting with our university president Prof. Rivka Carmi, our Rector, Prof. Zvi Hacohen, and our university Spokesman, Mr. Amir Rozenblit - Presented our 2014 iGEM project and discussed the importance of iGEM for both the university and the students.
Meeting Prof. Yaakov Bar Tana, expert at the Metabolic Syndrome field - presented our Synthetic Biology solution and discussed diabetes and pre-diabetes according to current and new treatments.
Meeting with DR. Younes Abu Rabia, the first Bedouin doctor at the Negev and expert in the diabetes field – worked together to accomplish our Human Practice goals.
Meeting with Sari Abu Saluk, a 4th year Nursing student- Cooperated in producing our "Metabolic Ambassadors" seminar for Bedouin students.
Meeting with the Israeli Minister of Health, Mrs.Yael German; the Ministry of Health Chief Executive, Prof. Arnon Afek; and Chief Scientist of the Ministry, Prof. Avi Yisraeli. Presented our "Inner Doctor" treatment, the Human Practice strategy and discussed the policy of product subsidy.
Meeting with prof.Riad Agbaria – presented our Bedouin Campaign and created continuity for our "Metabolic Ambassador" event in the years to come.
Week 7:September 14th-September 20th
 Prev
Prev
Lab
Biobricks-
First trial - the DNA parts that were used in Biobricks preparation are: Adiponectin (pUC 57 Adiponectin), DsbA-L (pUC 57 DsbA-L), HSP70 promotor (pUC 57 HSP70), HSP70 promotor (with modifications) (pUC 57 HSP70 modified), UCP1 (pcDNA3.1 UCP1) and pSB1C3 backbone. The process included- transformation -> extraction and creating a glycerol stock-> restriction-> gel electrophoresis-> gel products extraction-> ligation-> transformation.
Details:
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1
The transformation was done according to the general transformation protocol.
At first, heat-shock transformation to chemically competent BH5? bacteria with the following DNA parts (separately):
- pUC 57 Adiponectin
- pUC 57 HSP70
- pUC 57 HSP70 modified
- pUC 57 DsbA-L
- pcDNA3.1 UCP1
The transformed bacteria were incubated for 24 hr at 37?c.
After 24 hr incubation, colonies from each plate were transferred to liquid medium and incubated for 24 hr at 37?c.
Plasmid DNA was extracted using a miniprep kit and concentrations were measured using nanodrop.
The plasmids were digested with restriction enzymes Pst1 and EcoR1, following general restriction protocol.
The digested DNA parts were tested by gel electroporation, following general gel electrophoresis protocol.
Results:
The wanted bands didn’t appear on the gel.
Conclusions:
Since the desired results didn’t appear on the gel, it was assumed that the restriction didn’t work. The reason is probably technical inexperience of using restriction enzymes and inappropriate pipetation and spin down absence.
Intelligent Medication
Goal: Third try of trying to silence eGFP in CT26 eGFP cells – this time we also made a positive control with anti eGFP siRNA from our lab. Additionally we simplified the silencing process – using our mechanism, if transfected with 2 parts of the RNA construct silencing will take place only in the presence of a specific mRNA. This time we used only one part (part B – hairpin) that is supposed to have a silencing effect without the presence of the specific mRNA. We also tested our samples for silencing using RT PCR and flow cytometry (Image Stream).
Details:
The changes concluded from former results:
- Added positive control and used only part B hairpin:
- Anti eGFP siRNA - 200 pmol
- scRNA part B hairpin - 200 pmol
- Seeded cells amount was doubled, back to 100,000 cells per well.
The rest of the protocol remained unchanged (general lipofectamin protocol).
Results:
Image stream:
scRNA part B (hairpin) only

Positive control - anti GFP siRNA

Negative control-

Representative image stream pictures:

RealTime PCR:
The housekeeping genes showed unstable expression and thus could not be used as reliable control, rendering the results of the entire experiments unreliable.
Conclusions:
- The mean intensity in the GFP positive population of the positive control (anti GFP siRNa) is lower by 28 % of that in the negative control. However, this sequence was tested before and resulted in much higher silencing rates. With this, and in addition to previous results, we assume that flow cytometry, the way we use it, tests silencing in the protein level. Therefore it is likely that the proteins (eGFP is known to be stable) in the sample did not have enough time to degrade. Therefore, it was decided that image stream is not efficient test for the scRNA experiments and the appropriate test would deal with mRNA levels.
- The treatment containing scRNa part B only (hairpin silencing sequence) didn't show any lower GFP intensity in the GFP positive population, concerning the negative control.
- It was assumed that as image stream Real Time PCR results: Since the housekeeping genes showed unstable expression no significant result could be drawn regarding neither gene expression nor gene silencing from the RealTime PCR this time.
Human Practice
18/9/14- "Inner Doctor" video launch
19/9/14- Producing "Metabolic Ambassador" - Diabetes Seminar for 40 Bedouin students in different medical professions.
Week 8: September 21th-September 27th
 Prev
Prev
Lab
Biobricks-
Trying again the experiment from September 7th.
Details:
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1
The transformation was done according to the general transformation protocol.
At first, heat-shock transformation to chemically competent BH5? bacteria with the following DNA parts (separately):
- pUC 57 Adiponectin
- pUC 57 HSP70
- pUC 57 HSP70 modified
- pUC 57 DsbA-L
- pcDNA3.1 UCP1
The transformed bacteria were incubated for 24 hr at 37?c.
After 24 hr incubation, colonies from each plate were transferred to liquid medium and incubated for 24 hr at 37?c.
Plasmid DNA was extracted using a miniprep kit and concentrations were measured using nanodrop.
The plasmids were digested with restriction enzymes Pst1 and EcoR1, following general restriction protocol.
The digested DNA parts were tested by gel electroporation, following general gel electrophoresis protocol.
Ligation products were transformed by heat-shock to chemically competent DH5? bacteria, plated on cmp LB agar plates and incubated for 24 hr at 37?c.
Results:
All parts were successfully restricted (unfortunately the picture taken was not in good quality).
No colonies grew on the plates.
Conclusions:
Since no colonies grew on the plates, but restriction went well, the problem might lie either in the ligation process or the competent cells. For the next try, a new competent cells stock was used.
Biobricking third try:
Details:
DNA parts and pSB1C3 backbone (from pSB1C3 RFP and pSB1C3 amilcp) were digested with restriction enzymes: Pst1 and EcoR1
The ligation and transformation was done according to the previous protocols.
Results:
All parts were successfully restricted but again, no colonies grew on the plates.
Conclusions:
Since no colonies grew on the plates even though different competent cells stock was used, and restriction went well, we assume that the competent cells are not the problem and for the next try, we would use a new DNA ligase.
Human Practice
21/9/14- Meeting with a leading Consulting firm – 'Trigger Foresight' (Deloitte) - discussed the business aspect of the project and how it can affect the market.
Meeting with Huda Abu Obaid CEO of 'Yasmin' association (promoting health among Bedouin women) - presented our Human Practice events at the Bedouin community.
Week 9: September 28th- October 4th
 Prev
Prev
Lab
Biobricks
Forth trial, first for this week – With a new ligase enzyme, the rest of the process is identical to the last times.
Details:
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1
The transformation was done according to the general transformation protocol.
Results:
Restriction gel:


No colonies grew on the plates.
Conclusions:
Since still unsuccessful. We will try a different kit for purifying restriction products.
Fifth trial, second for this week
Details:
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1
The transformation was done according to the general transformation protocol.
Results:
All parts were successfully restricted, but again no colonies grew on the plates.
Conclusions:
Since a lot of changes was already done, and the time became critical for iGEM deadline, it was decided to try looking for a different lab and get some help.
Sixth trial, third for this week – with the former changes, we moved to Prof. Lital Alfonta's lab, using the lab members' knowledge and experience. The transformation was changed from Heat-Shock transformation to electroporation transformation.
Details:
DNA parts and backbone were digested with restriction enzymes: Pst1 and EcoR1
The transformation was done according to the general transformation protocol.
The gel products extraction, ligation and transformation was done at Prof. Lital Alfonta's lab with instruction of her lab members.
Results:
All parts were successfully restricted, electroporation time contants were all above 5 ms, but again no colonies grew on the plates.
Conclusions:
Human Practice
30/9/14- Producing a healthy cooking workshop led by Aaron Sulima, and early detection glucose tests for Bedouins spouses.
October
 Prev
Prev
General
*****
Week 10: October 5th- October 11th
 Prev
Prev
Lab
Biobricks-
Seventh and last trial, first for this week – with the former changes, working in different lab the insert vector ratio has been changed.
Details:
The transformation was done according to the general transformation protocol.
Protocol changes:
- Transformation by electroporation.
- The gel products extraction, ligation and transformation was done at Prof. Lital Alfonta's lab with instruction of her lab members.
- This time, the insert to vector ratio was raised to 4:1 instead of 3:1
Results:
The restriction worked, but again- No colonies grew on the plates.
Conclusions:
Since it was the last chance for preparing and sending the Biobricks to iGEM, unfortunately we have no more time to try and make this work. We suspect that step that prevented successful transformation was the ligation.
Intelligent Medication
Goal: Fourth try of trying to silence eGFP in CT26 eGFP – we repeated the exact protocol form the third try, but this time we will only try to analyze results by real time PCR.
Details:
Same as in third try.
Results:
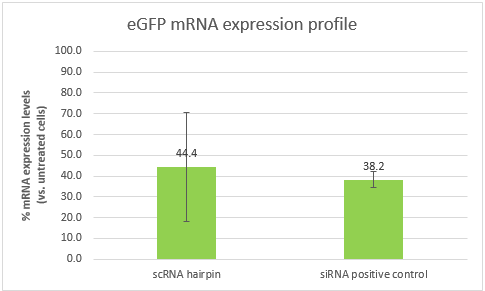
The results were analyzed USING THE ??CT method. The cells transfected with our scRNA hairpin contained only 44.4% eGFP mRNA in comparison to untreated cells (with a high standard deviation of 26.1%), while cells transfected with our positive control siRNA contained 38.2% mRNA.
Conclusions:The ability of our scRNA hairpin to perform silencing when it is unbound by the detection part of our construct is of crucial importance. These results show that it can silence a gene of interest, and in a similar efficiency to that of our positive control. However, the high standard deviation, will require us to repeat the experiment. It is also important to note that the efficiency of both samples is affected by the transfection efficiency, which is dependent of the transfection reagent. Optimizing the transfection might lead to better results and higher silencing efficiencies.
Human Practice
Meeting with the Mayor of Be'er Sheva (where our university is located), Mr Ruvik Danilovich , and his PR assistance, Amos Shavit - presented our project and the iGEM competition.
Week 11: October 12th-October18th
 Prev
Prev
Lab
Artificial Exercise
Goal: Use TMRM (tetramethylrhodamine methyl ester) to assess the ability of UCP1 to collapse membrane potential. Membrane potential-driven accumulation of TMRM within the inner membrane region of healthy functioning mitochondria results in a dramatic increase in TMRM-associated orange fluorescence. When the mitochondrial membrane potential collapses TMRM is dispersed throughout the cell cytosol at a concentration that yields minimal fluorescence upon excitation in the optimal wavelength region.
Details:
HepG2 cells were seeded in a 24 well plate (75,000 cells per well) and incubated for 24 hours at 37?c.
After 24 hours, in 3 of the wells the cells were transfected with pcDNA3.1 UCP1 using Lipofectamine, and incubated for 24 hours at 37?c.
Then, the cells were dyed with TMRM:
- The overnight medium was aspirated.
- Three different concentrations of TMRM 10 µM were added to 4 wells without any treatment: 0 nM, 10 nM, 25 nM, 50 nM, by adding 1 ml of medium DMEM with 0 µl, 1 µl, 2.5 µl, 5 µl of stock solution respectively .
- Incubate for 30 min in 37?C.
- The medium was replaced to DMEM without phenol red, 0.5 ml in each well.
- Same treatment was done with 3 wells of pcDNA3.1 UCP1 transfected HepG2 cells.
- The cells were viewed in a fluorescent microscope (excitation – 545 nm, emission – 575 nm).
Results:
The left series of pictures shows HepG2 cells treated with TMRM of different concentrations (left – white light, right – fluorescent), and the right series shows HepG2 previously transfected with pcDNA3.1 UCP1 as described above, also treated with TMRM of different concentrations.
There appears to be no visible difference between cells transfected with pcDNA3.1 UCP1 and the normal cell line. Also there was no difference between the different concentrations of TMRM, so next time we can use the lowest one (10 nM) or perhaps even lower.
Conclusions:
Human Practice
No Activity
Week 12
 Prev
Prev
Lab
No Activity
Human Practice
No Activity
Week 13
 Prev
Prev
Lab
No Activity
Human Practice
No Activity
 "
"

