Team:Austin Texas
From 2014.igem.org
| Line 87: | Line 87: | ||
Amino acids not included in the native 20 are commonly known as noncanonical amino acids (ncAAs). These ncAAs have unique chemistry that can provide very useful functionality not normally present in an organism. For our purposes, ncAAs were used to prevent protein functionality until spatially and/or temporally triggered. | Amino acids not included in the native 20 are commonly known as noncanonical amino acids (ncAAs). These ncAAs have unique chemistry that can provide very useful functionality not normally present in an organism. For our purposes, ncAAs were used to prevent protein functionality until spatially and/or temporally triggered. | ||
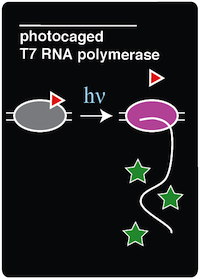
| - | This summer our team has been working on recreating a light-activatable T7 RNA Polymerase (RNAP) for a method of noninvasive, spaciotemporal control of protein expression. T7 RNAP was our enzyme of choice for this project due to the presence of a tyrosine residue in the active site of the polymerase. By | + | This summer our team has been working on recreating a light-activatable T7 RNA Polymerase (RNAP) for a method of noninvasive, spaciotemporal control of protein expression. T7 RNAP was our enzyme of choice for this project due to the presence of a tyrosine residue in the active site of the polymerase. By changing this codon to an amber codon, we were able to incorporate ortho-nitrobenzyl tyrosine (ONBY) into the active site, thus acting as a molecular cage. ONBY is decaged when exposed to 365 nm light. Thus, T7 RNAP is only functional when exposed to this light, which results in cleavage of the molecular cage from the polymerase’s active site. In our experiments, our photocaged ncAA (ONBY), once cleaved off, becomes normal tyrosine, restoring the T7 polymerase active site that is necessary for proper function of the protein. Once decaged, the polymerase can transcribe sequences that are preceded by a T7 promoter. To test this system, we used a GFP reporter to analyze spaciotemporal specificity of photo-activation in living cells, growing in media. We also attempted to photocage GFP. However, while the signal seen in live cells with photocaged T7 RNAP was quite strong, we could not detect decaged GFP in living cells. This is likely due to the amplifying effect of decaging a single molecule of T7 RNAP realtive to de-caging a single molecule of GFP. |
|- valign="top" | |- valign="top" | ||
| | | | ||
Revision as of 14:38, 17 October 2014
| |||||||||||||||||||||||||||||||||||
 "
"