Team:Heidelberg/pages/Human Practice/igemathome
From 2014.igem.org
(→Insight into Community) |
(→Insight into Community) |
||
| Line 244: | Line 244: | ||
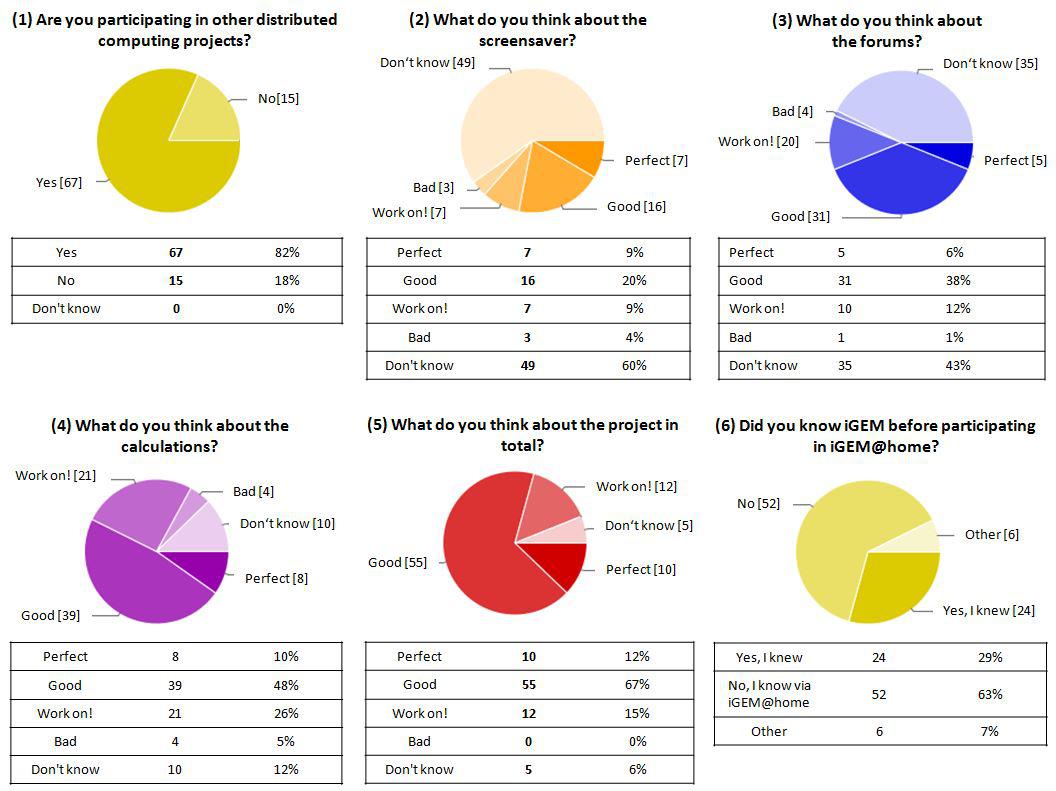
{{:Team:Heidelberg/templates/image-full|align=left|caption=What does our community think|descr=|file=iah_hp_piecharts.JPG}} | {{:Team:Heidelberg/templates/image-full|align=left|caption=What does our community think|descr=|file=iah_hp_piecharts.JPG}} | ||
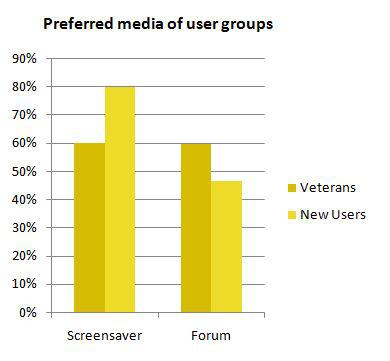
| - | {{:Team:Heidelberg/templates/image-quarter|align=right|caption=Veterans vs New Users|descr=Comparing the preferences of the BOINC veterans (already participating in projects) to the ones which are just calculating for | + | {{:Team:Heidelberg/templates/image-quarter|align=right|caption=Veterans vs New Users|descr=Comparing the preferences of the BOINC veterans (already participating in projects) to the ones which are just calculating for iGEM@home (New Users)|file=iah_hp_vet_vs_new.JPG}} |
=Survey and Meetup= | =Survey and Meetup= | ||
Revision as of 02:23, 17 October 2014
Contents |
Abstract(ReDo)
iGEM@home is a platform (build upon the BOINC framework) for the distribution of computations to volunteers who provide their computer resources when it is idle. The volunteers download a client software that pulls work from our central server and runs it when the computer is not used. After the successful execution of the computation the result is uploaded to our server. This approach allows future iGEM teams to run heavy computations without the need of access to a big server cluster. Furthermore the inclusion of general citizens in the project as providers for calculation power enables us to spread the word about Synthetic Biology circumventing the classical gatekeepers. During the development of the project we wanted to reach people who usually have nothing to do with science, let alone Synthetic Biology and iGEM. Our project offers a unique chance to not just tell people about Synthetic Biology, but to offer them the chance to participate and really get involved in a project. This will hopefully lead to new kinds of collaboration, better understanding and a broader acceptance of Synthetic Biology.
If you want to know more about the software that we developed please visit our Software Overview page, if you are rather interested in the modelling that we ran on iGEM@home and how it relates to our project please visit the Linker modeling.
Introduction
The creative concepts developed and the plethora of information gathered during the last decade by iGEM teams from all over the world proof efforts in Human Practices worth to be rewarded with a track of its own $Link in this year’s iGEM competition. Since iGEM teams from Heidelberg have always been interested in the question how laymen citizens can get involved into synthetic biology, they already introduced new concepts to the community to improve communication (first abstract page in multiple languages, first site tour for laymen, first mascot 2008), to do research on the mechanisms of science communication (2010) and to encourage reflection by philosophers and artists as representers of society (2010, 2013). Following the findings of our former team mates (https://static.igem.org/mediawiki/2010/7/72/Akzeptanz_end.pdf , also supported by literature as described in $Link) that acceptance towards a technology is not correlated with the information provided to a sceptic public, we rather aim at involving the public into the creative process of scientific work. In the style of the new iGEM community labs track that encourages science amateurs to contribute “beyond the accolades of scientific publishing and economic reward”, we sought for a new way to involve laymen in actual science and build a strong community of well informed supporters and communicators of synthetic biology at the same time. Thereby we want to lay the foundation for a democratization of Science in an enlightened society. Now we proudly present the crowdsourcing and communication platform iGEM@home!
Introducing Distributed Computing to the iGEM community
Modern biology uses computational simulations of models for validation as well as to predict key experiments and suitable designs for specific applications. Despite of the fact that the experience with and the mere amount of algorithms and code dedicated to biological systems is growing exponentially these days, big data evaluation such as sequence analysis and multidimensional problems such as protein folding still consume vast amounts of computational resources. Therefore main aspects of biological science are only accessible to research institutions that provide these resources in form of supercomputers or computer clusters. In order to open computational biology up to all future iGEM teams in a manner that is not dependant of institutional affiliations, we decided to develop a platform from a set of applications that build upon the BOINC framework for distributed computing $Link. Thereby we provide access to computing power for future (community lab) iGEM teams and at the same time create an opening for laymen to jump in and empower science! $Link
BOINC and iGEM - Foundations of a rewarding relationship
The BOINC framework distributes computational tasks to volunteers who provide computational resources in form of calculation time spent by the CPU of their idling personal computers. Volunteers download and install a client software that receives work packages from a central server unit and runs calculations once the computer is not in use. Alternatively users may dedicate a certain percentage of their computing power to destributed computing projects that they like to support. After the successful execution of the computation the result is uploaded to our server. One of the best known distributed computing projects is SETI@home (http://setiathome.ssl.berkeley.edu/), a project in which volunteers contribute computing power to the search for extra-terrestrial intellegance. In our case the need for computing power arose from the calculations needed to evaluate the linker model that provides rigid linkers for the circularization of proteins $LInk. In both cases the contributors are rewarded with points that reflect the computing time already spent and the dedication towards certain projects. In order to take motivation to the next level we implemented a platform that allows to feed information to the BOINC screensaver (figure 1). Thereby we enable iGEM teams from all over the world to reach an interested and motivated audience with information about their projects, Human Practices activities and iGEM and Synthetic Biology in general. In return users of iGEM@home will be rewarded with informations and insights into one of the most vital communities in science that catch their attention in the very moment they get back to their computers.
Outreach
The need to build a vivid community is key to run a successful distributed computing project. As the project depends on the voluntary support of people it is important to get people enthused about the science behind the project, so they are willing to support the project. Good communication is therefore essential to find support. We therefore used different channels to present and promote our project and iGEM in general to convince people to help us.
Promoting iGEM@home: Material and Methods
The homepage of iGEM@home
Website
A central part of was our campaign for iGEM@home is the website [http://www.igemathome.org www.igemathome.org] that is the frontend of our BOINC server and therefore needed to be externally hosted. (To find out more about the software click here). It bundles all important information on the [http://www.igemathome.org/project project] and explains iGEM in a layman’s way to an idea in what context this project was developed. It also provides a list of [http://www.igemathome.org/FAQ FAQ’s] that address most important concerns and questions that people have. In addition we also provide a list of [http://www.igemathome.org/related_projects other BOINC projects], because the client makes it very easy to support additional projects from different areas of science. Furthermore the website also hosts the [http://www.igemathome.org/igemathome/ “BOINC pages”]. These are the pages, where participants can view the server status, manage their account, create a profile and join the discussion in the [http://www.igemathome.org/igemathome/forum_index.php forums]. (For more information on the role of the forum in the outreach you can skip to #Forums.)
Video
The second pillar of our promotion is a video that provides the information in an easily digestible way.
The video presents iGEM, our project and distributed computing. It explains why we need the support of the users and how to help us.
Flyer
To promote iGEM@home on local level to friends, fellow students and people interested in science and iGEM we created Flyers that are supposed to make people interested in the project and lead them (via QR-code or link) to our website.




Media Campaign
Although iGEM@home is open for everyone to participate, we decided to promote the project mainly to people we knew (friends, family, fellow students etc.) and people that are already active in the BOINC community. We used social media and forums to present the project and shared our video as it is the easiest way to get informed. Moreover our project was covered by 2 journals ([http://www.spektrum.de/news/proteinforschung-zum-mitmachen/1307953 Spektrum der Wissenschaft] german popular science magazine and [http://www.spektrum.de/news/proteinforschung-zum-mitmachen/1307953 BioTechniques] an international journal on Life Science Methods) who wrote articles about the project, which was very helpful to reach new user groups.
The Communication Platform iGEM@HOME
The important job after convincing the user to try our software, is to convince them to continuously support us by constantly feeding them with interesting facts on iGEM and offering them ways to influence the project.
Forum: Feedback on Computing and Projects
The first pillar for the communication is an integrated [http://www.igemathome.org/igemathome/forum_index.php forum] software, where the user can help each other and directly contact as us the developers if they experience bugs.
Screen Saver: Communication Channel for Synthetic Biology
The second pillar is a [http://www.igemathome.org/screensaver screensaver], where we can show the users interesting facts on science and promote the projects [Link collaboration with Marburg] of fellow iGEM teams. This bidirectional communication enabled an optimal exchange between our users and us as the project developers.
Evaluation of iGEM@home, a new tool in Science Communication
(as of 12.10.2014)
Users
- 400 active users
- From 42 countries
- 3/4 supporting additional BOINC projects
- Organized in 80 teams
- Age ranging from 12 year old brother to 88 year old grandma
Computation power
- > 1,100 actively calculating computers
- > 100,000 computed jobs
- 1.2 Million CPU hours
*
- Equivalent to a 28,190$ per month cloud computer cluster by amazon web services
*
Communication
- 190 forum posts (mostly bug reports and ideas for next development steps)
- Showed screensaver 1300 times for an average of 2:08 hours
- Showed nearly 40,000 screensaver slides
For the most up to date information visit the [http://www.igemathome.org/igemathome/server_status.php server status page], the official [http://boincstats.com/en/stats/155/project/detail/overview BOINCstats] and our [http://igemathome.org/igemathome/forum_index.php forum].
Insight into Community
Survey and Meetup
Follows soon...
Conclusion
We consider iGEM@home a great success. Not just were we able to do calculations impossible without a huge server cluster, we also reached a lot of people who didn’t know about Synthetic Biology and iGEM before. The biggest part of our user base are people who are already contributing to BOINC projects, but this group is only united by their interest for number crunching and is in many other aspects quite divers.
We hereby have shown that it is feasible for an iGEM team to run an distributed computing project and it offers a great new approach to reach out to the society.
Future
Of course we think that iGEM@home is a great tool and needs to be continued after the end of this year’s iGEM competition. As it has an active and interested community and offers teams to do impressive computations, we hope to hand iGEM@home over to the headquarters for a centralized access in future. So that it becomes an integral part of iGEM, for the benefit of society.
 "
"