Team:Austin Texas/photocage
From 2014.igem.org
Nathanshin (Talk | contribs) (→Results) |
Nathanshin (Talk | contribs) (→Experimental Methods) |
||
| Line 112: | Line 112: | ||
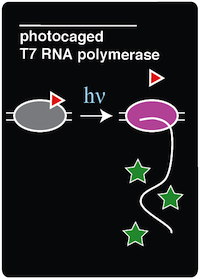
| - | In order to create an in vivo light-activated GFP expression system, two plasmids were constructed and transformed into amberless E.coli cells. The first plasmid contains the tRNA and synthetase pair, which is necessary to incorporate ONBY into the amber stop codon of the T7 RNAP. The second plasmid contains the coding sequence for T7 RNAP which has a mutation on the Tyrosine 639 residue | + | In order to create an in vivo light-activated GFP expression system, two plasmids were constructed and transformed into amberless E.coli cells. The first plasmid contains the tRNA and synthetase pair, which is necessary to incorporate ONBY into the amber stop codon of the T7 RNAP. The second plasmid contains the coding sequence for T7 RNAP, which has a mutation on the Tyrosine 639 residue, and a GFP coding sequence bound to an upstream T7 promoter. Once these components were assembled by Gibson Assembly, the two plasmids were transformed via electroporation into aliquots of Amberless E.coli. |
| - | + | ||
| - | Once these components were assembled by Gibson Assembly, the two plasmids were transformed via electroporation into aliquots of Amberless E.coli. | + | |
After transformation, the amberless E. coli cells containing both the synthetase/tRNA pair plasmid and the amber T7 RNAP/GFP plasmid were assayed for their ability to express GFP upon irradiation with 365 nm light in different intervals of time. The cells were irradiated for 0 minutes, 1 minute, 5 minutes, 15 minutes, and 30 minutes in order to observe the temporal control of GFP expression. | After transformation, the amberless E. coli cells containing both the synthetase/tRNA pair plasmid and the amber T7 RNAP/GFP plasmid were assayed for their ability to express GFP upon irradiation with 365 nm light in different intervals of time. The cells were irradiated for 0 minutes, 1 minute, 5 minutes, 15 minutes, and 30 minutes in order to observe the temporal control of GFP expression. | ||
| + | |||
| + | For this experiment, there were also other necessary control strains to test alongside the experimental strain. These controls included a T7-GFP construct with no amber stop codon in the O-helix (to serve as a positive control for expression with T7 polymerase), sfGFP amberless E.coli (to observe the expression of GFP by native polymerase), amberless E.coli (to serve as a cell background control), and LB supplemented with ncAA (to serve as a media background control). | ||
| + | |||
After irradiation with light, these cells were allowed to grow overnight before taking fluorescent measurements. This additional growth was necessary to allow the "decaged" T7 RNAP to polymerize mRNA transcripts of the GFP coding sequence. | After irradiation with light, these cells were allowed to grow overnight before taking fluorescent measurements. This additional growth was necessary to allow the "decaged" T7 RNAP to polymerize mRNA transcripts of the GFP coding sequence. | ||
Revision as of 01:58, 17 October 2014
| |||||||||||||||||||||||||||||
 "
"