Team:Austin Texas/kit
From 2014.igem.org
(→References) |
(→Motivation) |
||
| Line 86: | Line 86: | ||
'''Desired properties??? High fidelity? High efficiency of incorporation? We should indicate what we want our kit to test for.''' | '''Desired properties??? High fidelity? High efficiency of incorporation? We should indicate what we want our kit to test for.''' | ||
==Motivation== | ==Motivation== | ||
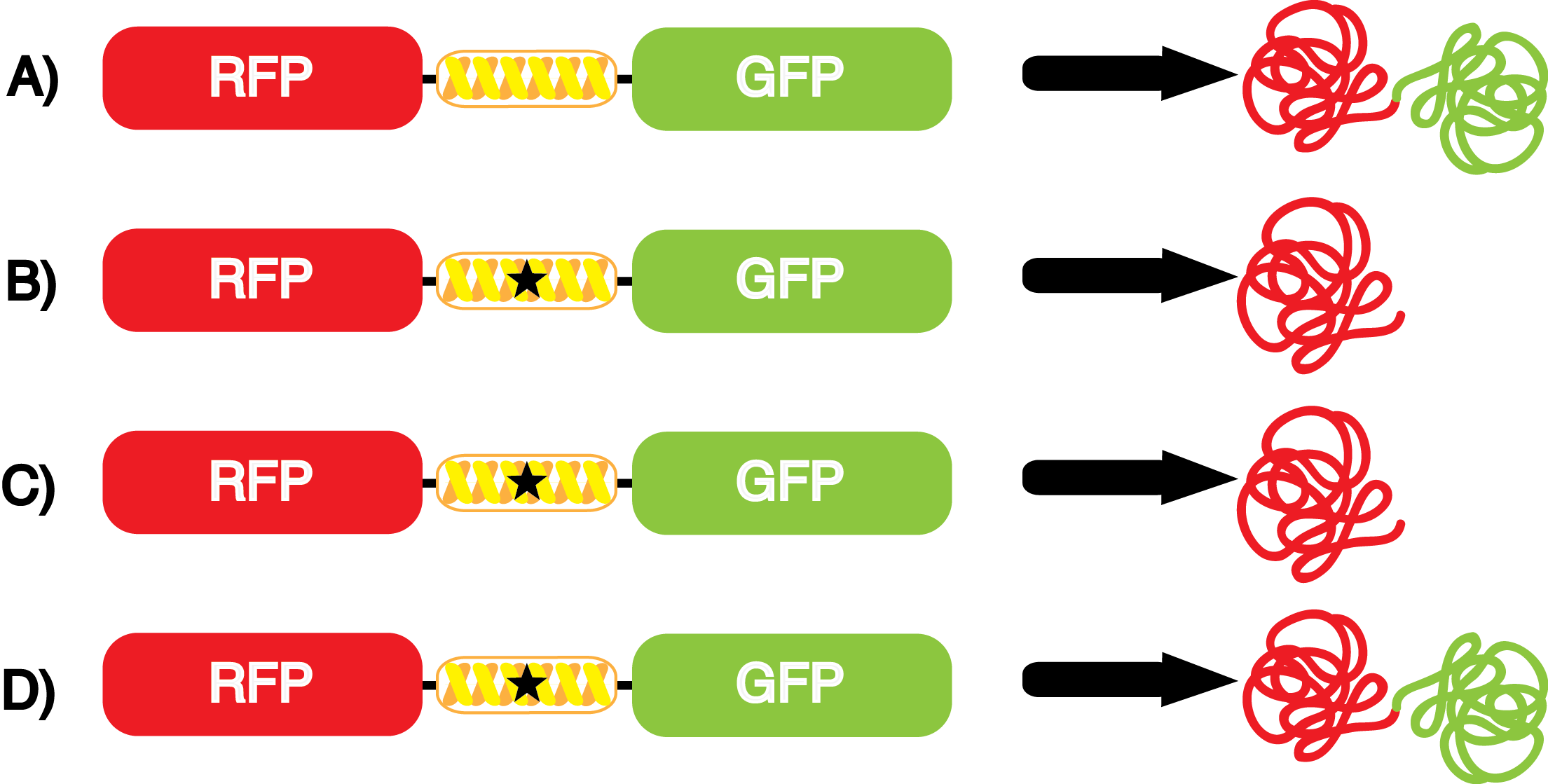
| - | In order to recode UAG, a synthetase must be mutated to effectively "charge" a ncAA onto the corresponding tRNA '''FIGURE?'''. Various methods of directed evolution | + | In order to recode UAG, a synthetase must be mutated to effectively "charge" a ncAA onto the corresponding tRNA '''FIGURE?'''. Various methods of directed evolution are typically used to modify a synthetase such that it can interact with and then charge a specific ncAA (Liu et al. 2010). The ncAA synthetases available have ranging levels of reported efficiency and are not well characterized. Many of the ncAA are not widely used, they are published in short articles lacking full documentation, and our own unpublished experiences of working with a number of ncAAs indicate that not all ncAA synthetases are created equal. Thus, we created a standard kit designed to characterize the properties of any ncAA synthetase/tRNA pair. Our goal is to produce a standardized kit that is cheap, reproducible, easy to use, AND easily portable (i.e. you don't need a lot of advanced equipment). |
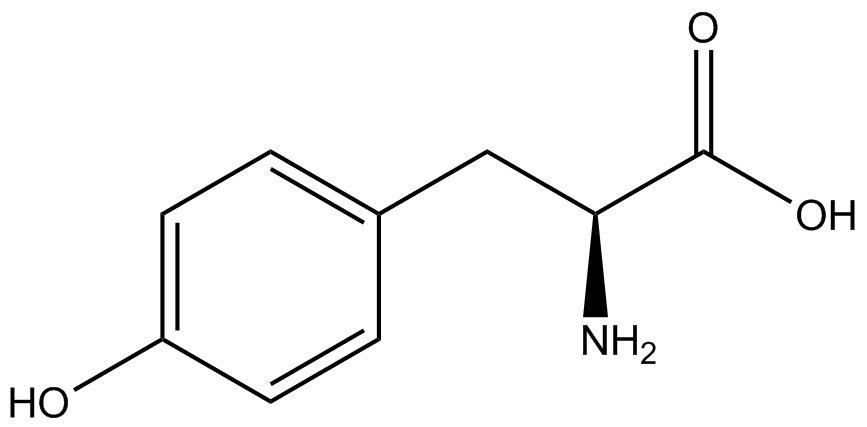
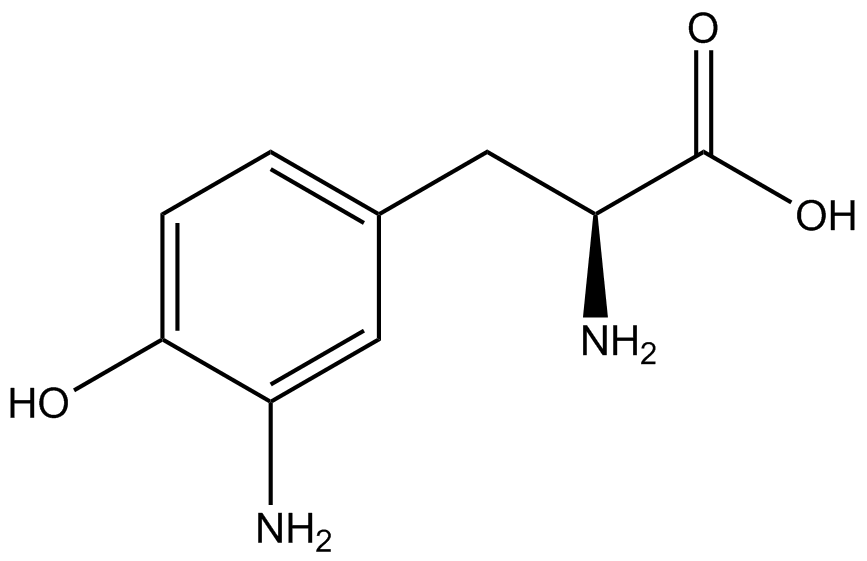
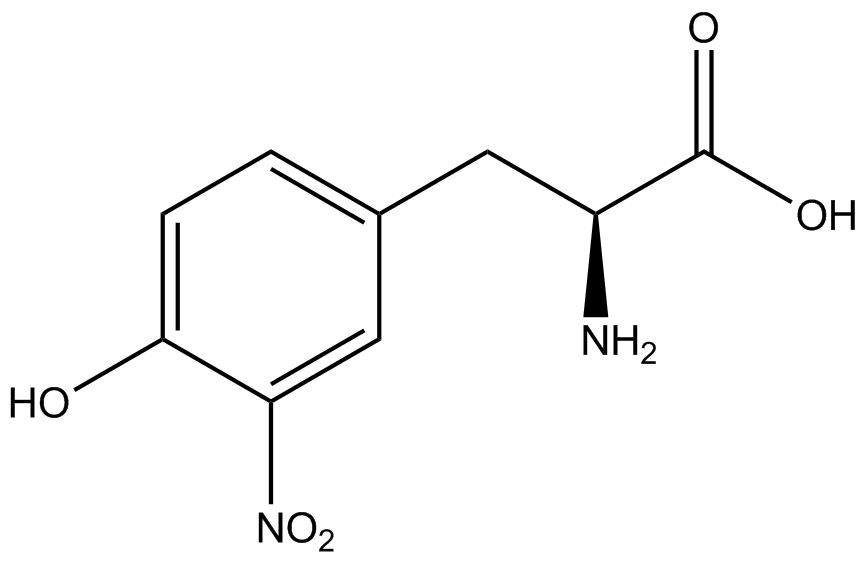
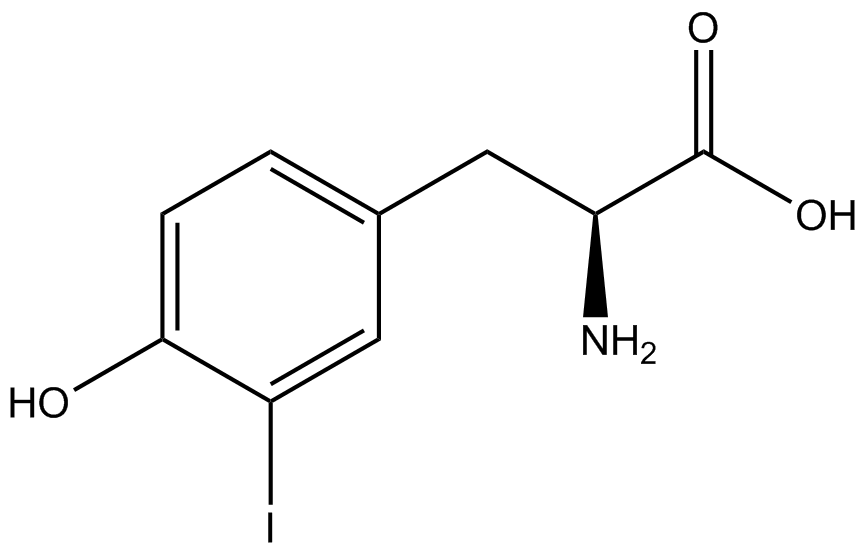
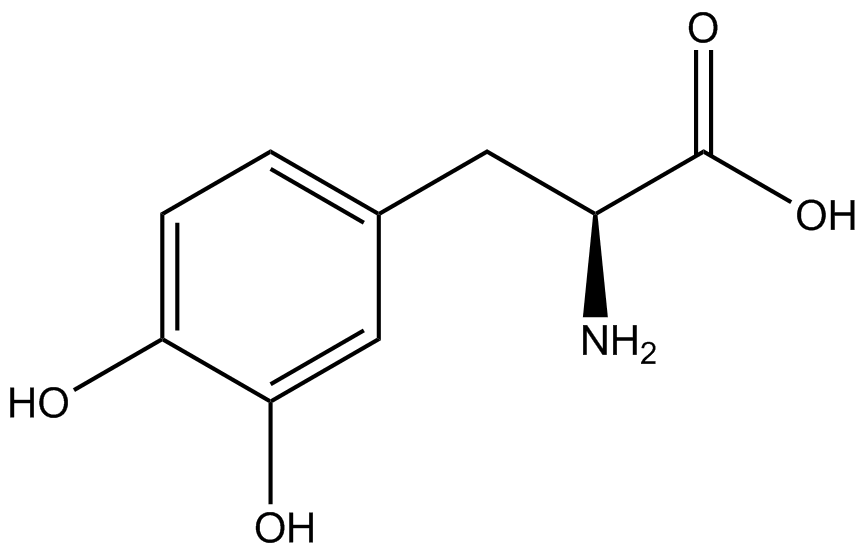
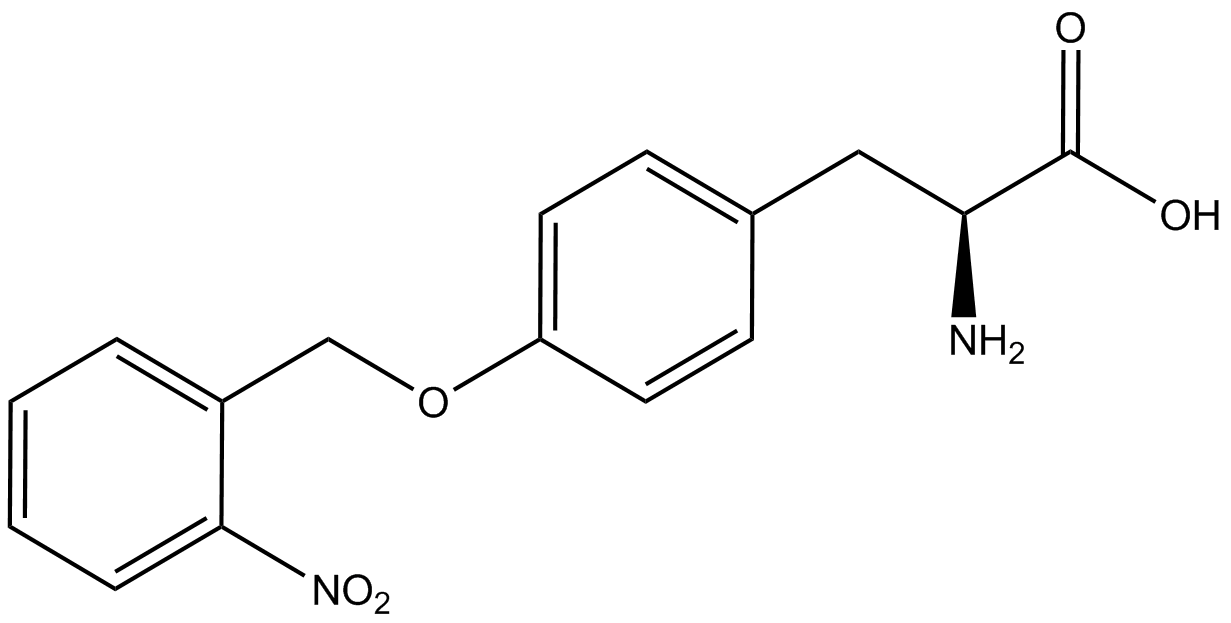
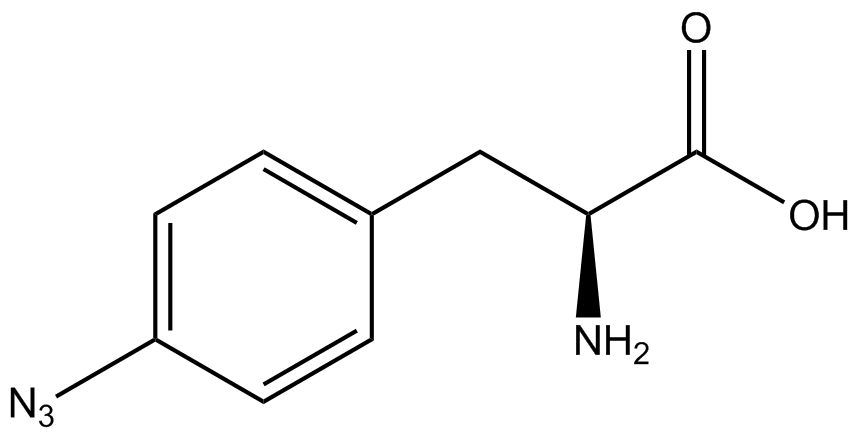
A total of seven ncAAs were used in addition to tyrosine. [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table The full list of amino acids used in this study can be found here]. | A total of seven ncAAs were used in addition to tyrosine. [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table The full list of amino acids used in this study can be found here]. | ||
Revision as of 00:11, 17 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"