Team:Austin Texas/kit
From 2014.igem.org
(→Fidelity of Incorporation) |
|||
| Line 185: | Line 185: | ||
[[File:UT_Austin_2014_Kit_Normalized_GFP_to_RFP_graph.png|thumb|600px|Figure 3. Graph showing the level of GFP fluorescence relative to RFP fluorescence. Each pStG plasmid is referred to based upon the tRNA synthetase/tRNA pair present in the specific plasmid. Each of these plasmids was then paired with either pFRY or pFRYC and grown in the presence or absence of a specific ncAA. For example, the "3-AminoY-FRYC" and the "3-AminoY-FRY" samples both contain the 3-AminoY synthetase/tRNA pair and both samples were grown in the absence or presence of the ncAA "3-AminoY". Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] | [[File:UT_Austin_2014_Kit_Normalized_GFP_to_RFP_graph.png|thumb|600px|Figure 3. Graph showing the level of GFP fluorescence relative to RFP fluorescence. Each pStG plasmid is referred to based upon the tRNA synthetase/tRNA pair present in the specific plasmid. Each of these plasmids was then paired with either pFRY or pFRYC and grown in the presence or absence of a specific ncAA. For example, the "3-AminoY-FRYC" and the "3-AminoY-FRY" samples both contain the 3-AminoY synthetase/tRNA pair and both samples were grown in the absence or presence of the ncAA "3-AminoY". Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] | ||
| + | '''Set the experiment up - What do we want to test? How are we going to test it (keep it simple)?''' | ||
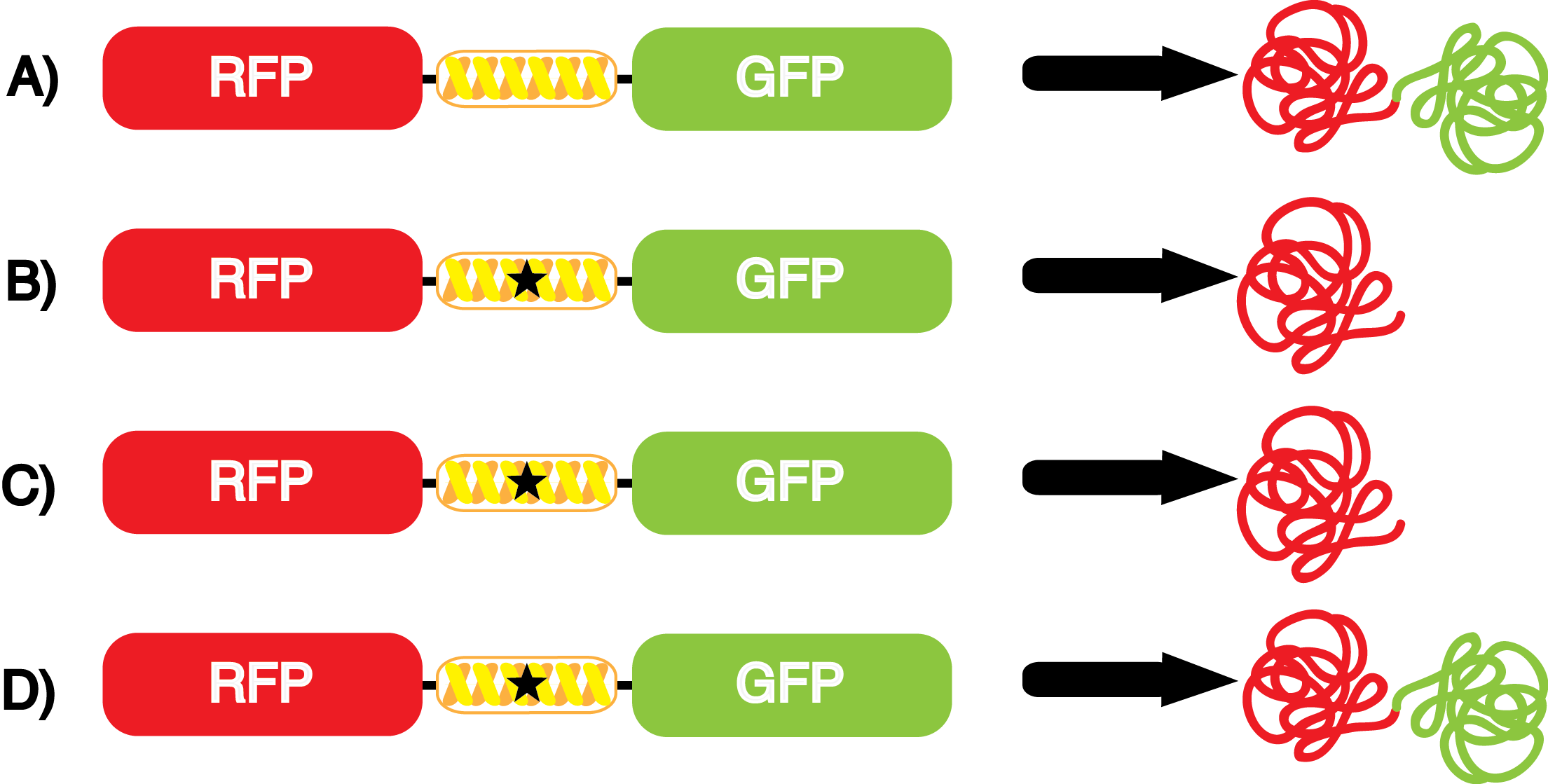
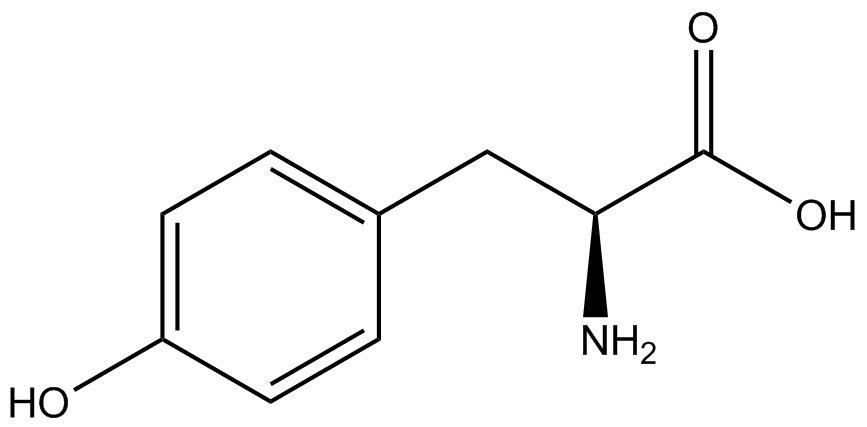
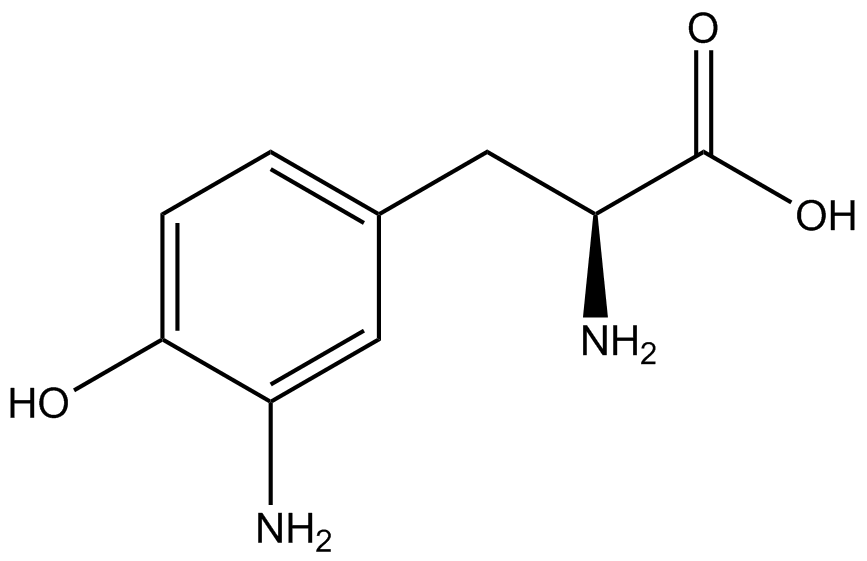
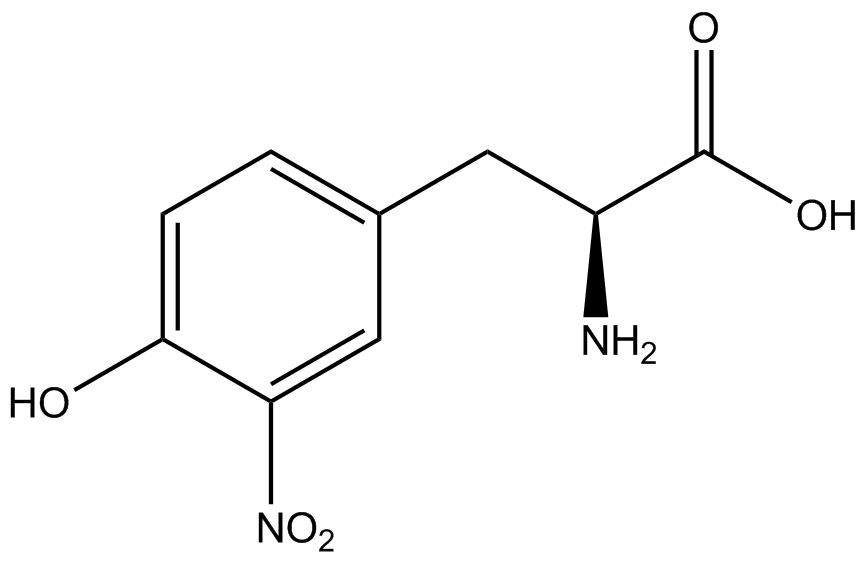
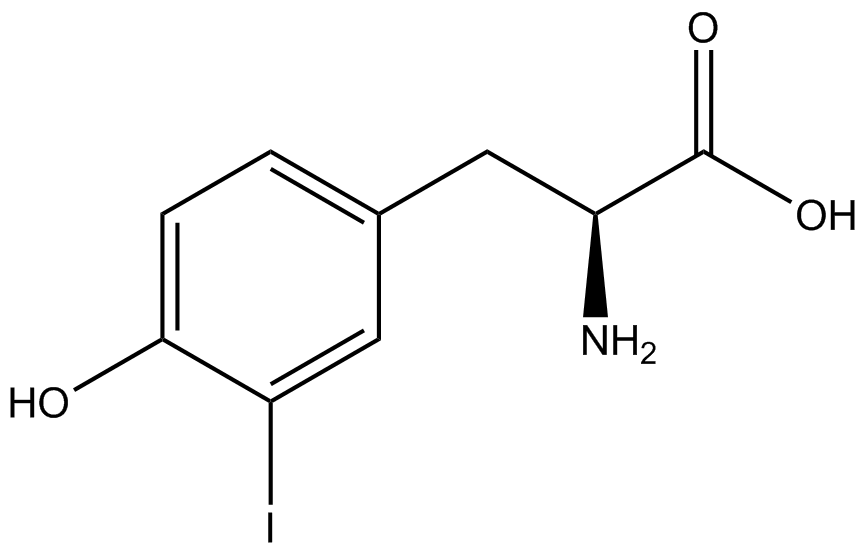
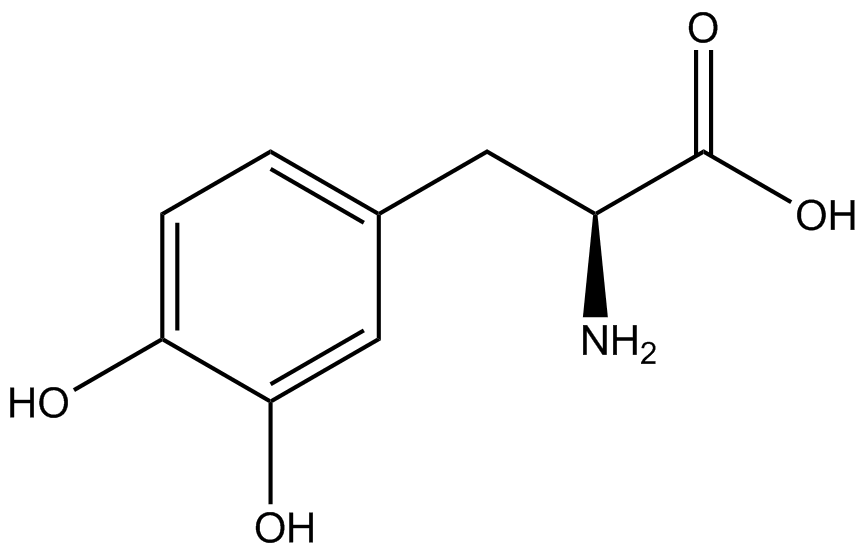
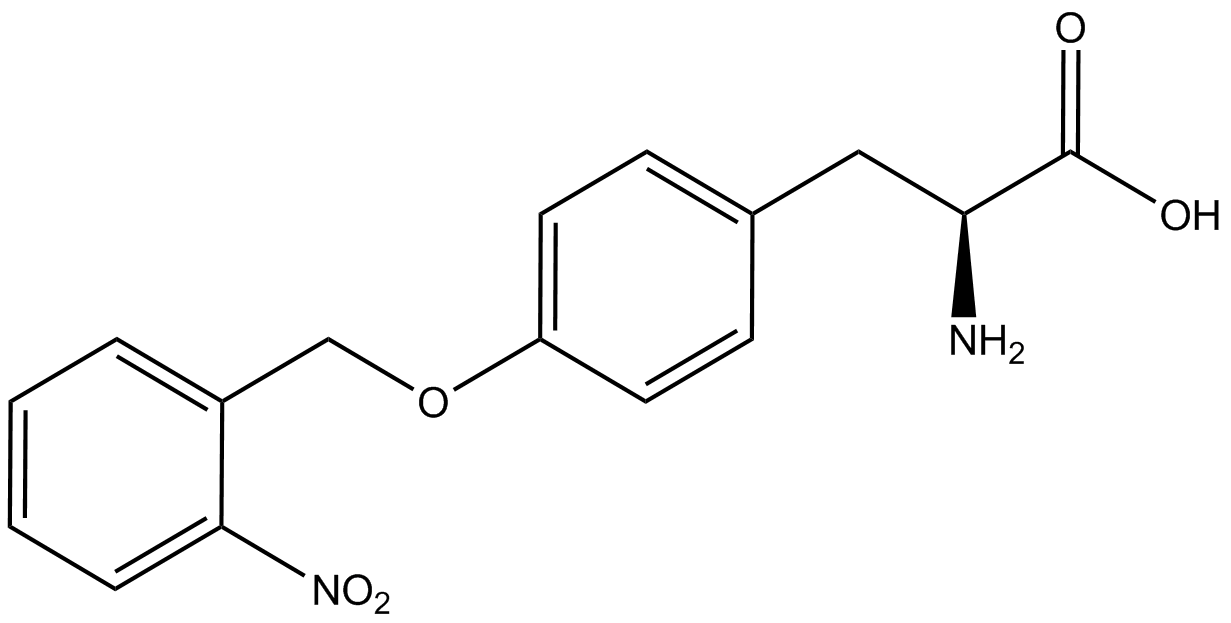
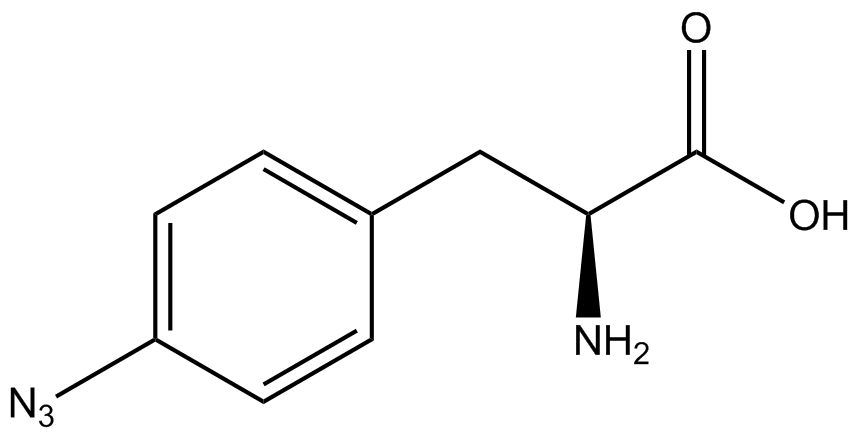
The fidelity of each ncAA sythetase/tRNA pair was measured by comparing the GFP production of pStG/pFRY strains in (+/-) ncAA conditions. In the absence of a non-canonical only RFP should be translated, as translation is expected to terminate between RFP and GFP at the amber stop codon (UAG) on the linker sequence in pFRY. Alternatively, if the corresponding non-canonical amino acid for pStG is present or if the synthetase/tRNA pair is not specific enough, translation should continue through the linker due to its incorporation at UAG. In this case, RFP and GFP should both be translated. The amount of fluorescence from RFP and GFP is detectable by fluorometer. We also tested pStG/pFRYC strains in (+/-) ncAA conditions as a control, which should express both RFP and GFP no matter what due to the lack of the amber stop codon in the linker (with a tyrosine in its place). Without the stop codon, there would be no reason to halt translation before the GFP. | The fidelity of each ncAA sythetase/tRNA pair was measured by comparing the GFP production of pStG/pFRY strains in (+/-) ncAA conditions. In the absence of a non-canonical only RFP should be translated, as translation is expected to terminate between RFP and GFP at the amber stop codon (UAG) on the linker sequence in pFRY. Alternatively, if the corresponding non-canonical amino acid for pStG is present or if the synthetase/tRNA pair is not specific enough, translation should continue through the linker due to its incorporation at UAG. In this case, RFP and GFP should both be translated. The amount of fluorescence from RFP and GFP is detectable by fluorometer. We also tested pStG/pFRYC strains in (+/-) ncAA conditions as a control, which should express both RFP and GFP no matter what due to the lack of the amber stop codon in the linker (with a tyrosine in its place). Without the stop codon, there would be no reason to halt translation before the GFP. | ||
Revision as of 20:46, 16 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"