Team:Austin Texas/kit
From 2014.igem.org
(→Experimental Design and Method) |
(→Using The Kit) |
||
| Line 137: | Line 137: | ||
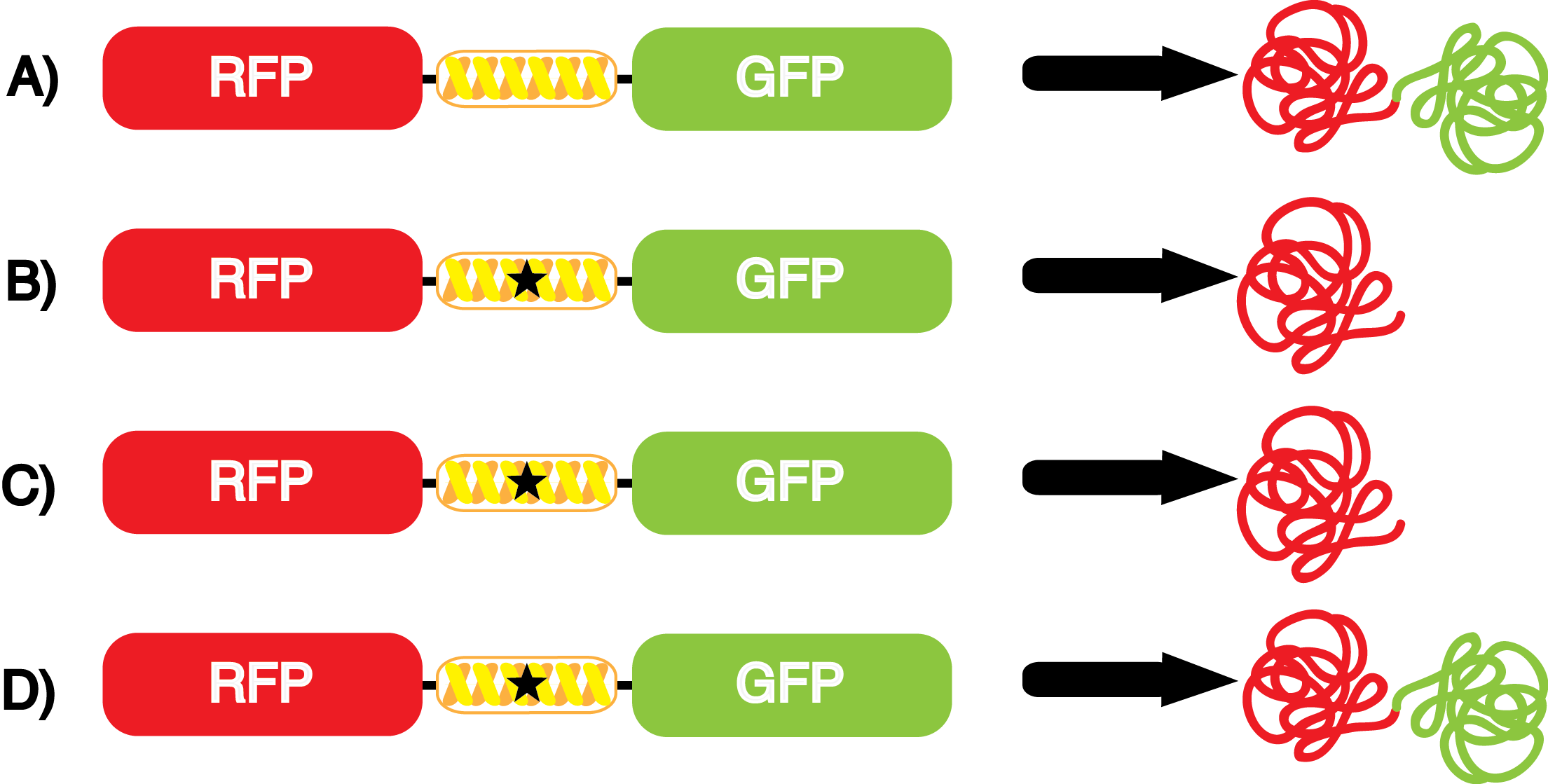
However, the above scenarios assume that the synthetase/tRNA pair function efficiently and with high fidelity. | However, the above scenarios assume that the synthetase/tRNA pair function efficiently and with high fidelity. | ||
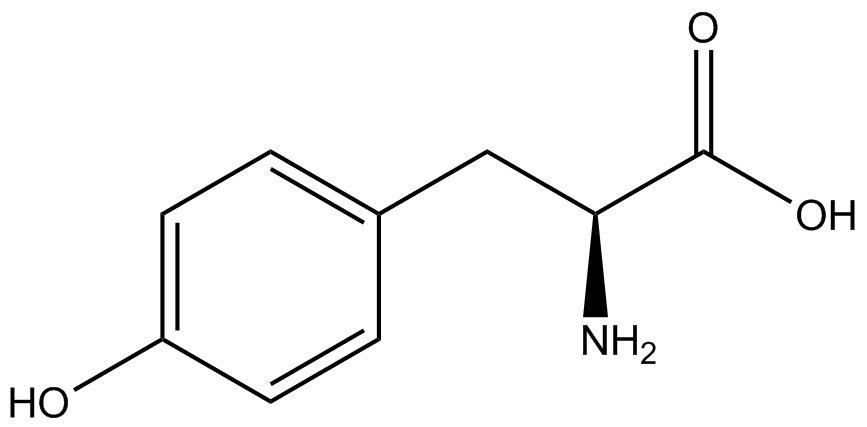
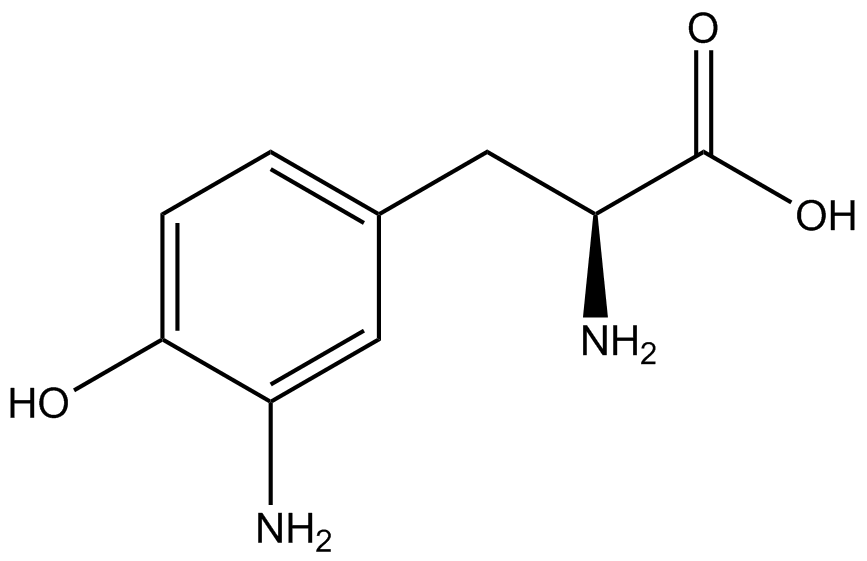
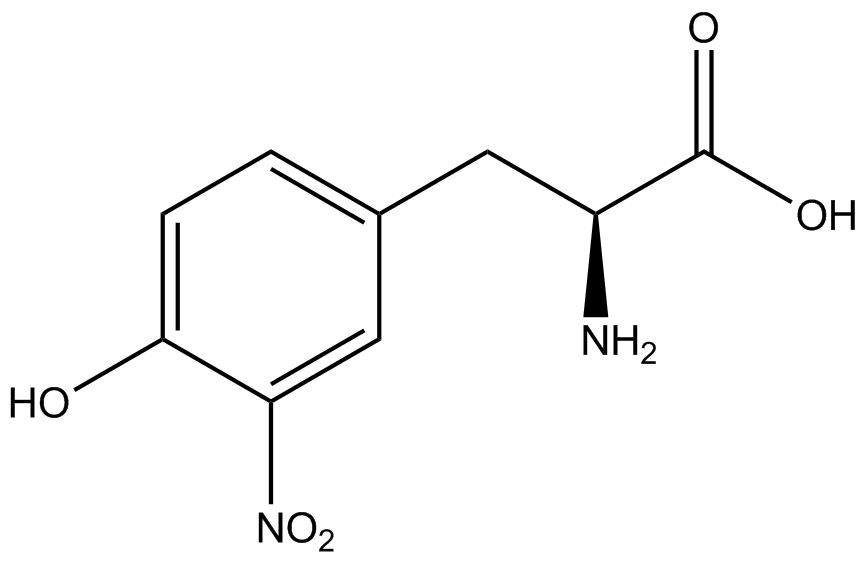
| - | *Actual synthetase/tRNA pairs sometimes have '''low fidelity''', leading to the incorporation of an incorrect amino acid. | + | *Actual synthetase/tRNA pairs sometimes have '''low fidelity''', leading to the incorporation of an incorrect amino acid. This "misincorporation" means that such a synthetase/tRNA pair would not give results that mimic Figure 2C in the absence of ncAA. Rather, in the absence of ncAA, such a pair would give results similar to Figure 2D, full expression of RFP and sfGFP. High rates of misincorporation are equivalent to '''low fidelity''', and tRNA synthetases with low fidelity are not ideal. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | *Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in Figure 2D might be significantly lower than expected. '''low efficiency''' is also not ideal as it may limit levels of protein expression. | ||
==Experimental Preparation== | ==Experimental Preparation== | ||
Revision as of 20:15, 16 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"