Team:ETH Zurich/modeling/xor
From 2014.igem.org
(→XOR biologic gate) |
(→XOR biologic gate) |
||
| Line 34: | Line 34: | ||
=== XOR bio''logic'' gate === | === XOR bio''logic'' gate === | ||
| - | [[File:ETHZ_XORmodule.png|center|600px|The behaviour of XOR module as a function of activated Bxb1 sites (SABxb1) and ΦC31 sites (SAΦC31). The XOR behaviour is continuous since we modelled it deterministically.]] | + | [[File:ETHZ_XORmodule.png|center|600px|thumb|The behaviour of XOR module as a function of activated Bxb1 sites (SABxb1) and ΦC31 sites (SAΦC31). The XOR behaviour is continuous since we modelled it deterministically.]] |
Revision as of 08:46, 14 October 2014
XOR Gate
Model
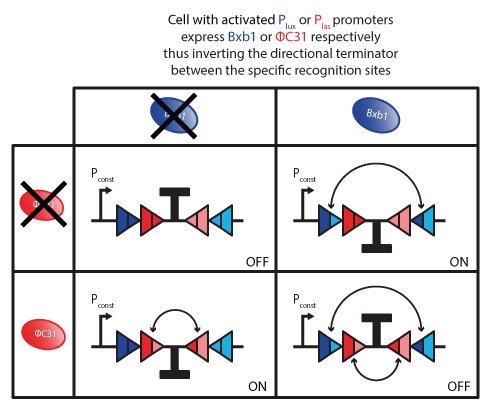
After binding to DNA, integrases can flip the fragment and thus compute the output of the XOR logic gate.
XOR Logic Gate
The truth table of an XOR gate in binary
Biological Principles
The fragment integrases can flip is a terminator. Thus, the terminator can either be on or off.
- Ton: terminator is on, transcription is blocked.
- Toff: terminator is off, transcription is active. It corresponds to one flipping of the terminator.
In our design, we are interested in a double flipping. That is to say that two pairs of binding sites surrounds the fragment to be flipped. One pair of binding sites can be bound by DBxb1 and the other one by ΦC31.
The terminator can be flipped once if either DBxb1 or ΦC31 is present. The state Toff can be caused by two reasons. We further decompose it into two different states: ToffBxb1(flipping due to presence of Bxb1) and ToffΦC31(flipping due to presence of ΦC31).
Insert Image of two states
Flipping by integrases is irreversible. The initial state, in which the terminator is on, is different from the state after two switches. From this last state, no further evolution of the system is possible. Therefore, we decompose the Ton into two different states: Ton,i(initial state of the system, no flipping) and Ton,f(final state of the system after two flips).
Insert Image of two states
XOR biologic gate
Other Chemical Species
| Name | Description |
|---|---|
| mRNAGFP | mRNA for Green fluorescent protein which is produced when the cells are ON. |
| GFP | Green fluorescent protein which is produced when the cells are ON. |
| mRNALuxI | mRNA for LuxI which is produced when the cells are ON. |
| LuxI | Enzyme catalysing the production of LuxAHL from SAM and ACP. |
| mRNALasI | mRNA for LasI which is produced when the cell are ON. |
| LasI | Enzyme catalysing the production of LasAHL from SAM and ACP. |
Reactions
The following reactions are valid for the strain producing LasAHL as output (It corresponds to the blue cells here). To have the equivalent for the strain producing LuxAHL as output, it suffices to remplace every occurence of LasI by LuxI. $$\begin{align*} SA_{Bxb1}+SA_{Bxb1}+T_{on,i}& \rightarrow T_{offBxb1}+ SF_{Bxb1}+SF_{Bxb1}\\ T_{offBxb1} &\rightarrow T_{offBxb1} + mRNA_{GFP} + mRNA_{LasI} \\ SA_{\phi C31}+SA_{\phi C31}+T_{on,i}& \rightarrow T_{off\phi C31}+SF_{\phi C31}+SF_{\phi C31}\\ T_{off\phi C31} &\rightarrow T_{off\phi C31} + mRNA_{GFP} + mRNA_{LasI} \\ SA_{Bxb1}+SA_{Bxb1}+T_{off\phi C31}& \rightarrow T_{on,f}+SF_{Bxb1}+SF_{Bxb1}\\ SA_{\phi C31}+SA_{\phi C31}+T_{offBxb1}& \rightarrow T_{on,f}+SF_{\phi C31}+SF_{\phi C31}\\ mRNA_{GFP} &\rightarrow GFP\\ mRNA_{LasI} &\rightarrow LasI\\ mRNA_{GFP} &\rightarrow \emptyset\\ mRNA_{LasI}&\rightarrow \emptyset\\ GFP &\rightarrow \emptyset\\ LasI&\rightarrow \emptyset \end{align*}$$
 "
"