Team:Austin Texas/photocage
From 2014.igem.org
(→Results) |
(→Background) |
||
| Line 95: | Line 95: | ||
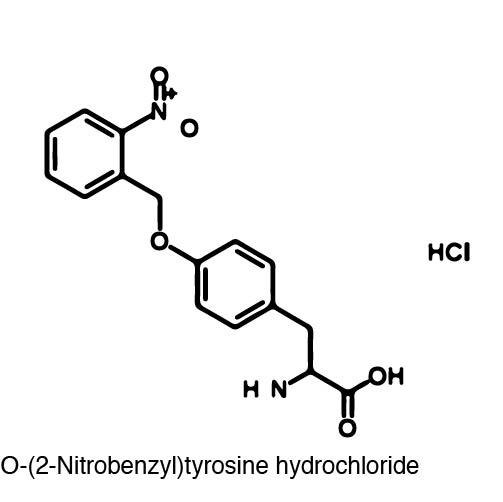
Tyrosine residue 639 (Y639) was specifically targeted because it lies on a crucial position on the O-helix and has been proved to be essential for polymerization. (REFERENCE) The Y639 residue in the O-helix is responsible for two major roles. First, this tyrosine residue discriminates between deoxyribose and ribose substrates. Second, Y639 is responsible for moving newly synthesized RNA out of the catalytic site and preparing for the next NTP to be inserted. These functions of the O-helix were shown to be essential through mutational analysis. Because the loss of this tyrosine residue in the active site leads to a non-functional polymerase, Y639 proved to be a good candidate for incorporating a photocaged amino acid (Reference; You are referring to a specific result). | Tyrosine residue 639 (Y639) was specifically targeted because it lies on a crucial position on the O-helix and has been proved to be essential for polymerization. (REFERENCE) The Y639 residue in the O-helix is responsible for two major roles. First, this tyrosine residue discriminates between deoxyribose and ribose substrates. Second, Y639 is responsible for moving newly synthesized RNA out of the catalytic site and preparing for the next NTP to be inserted. These functions of the O-helix were shown to be essential through mutational analysis. Because the loss of this tyrosine residue in the active site leads to a non-functional polymerase, Y639 proved to be a good candidate for incorporating a photocaged amino acid (Reference; You are referring to a specific result). | ||
| - | [[Image:Uncaging_of_ONBY.jpg | 300px|left|thumb| Figure 2. The caged T7 RNAP is decaged via exposure to 365 nm light.]] | + | [[Image:Uncaging_of_ONBY.jpg | 300px|left|thumb| Figure 2. The caged T7 RNAP is decaged via exposure to 365 nm light. '''SOURCE OF IMAGE? NEED REFERENCE''']] |
Revision as of 21:29, 13 October 2014
|
 "
"