Team:Valencia UPV/Modeling/fba
From 2014.igem.org
Alejovigno (Talk | contribs) |
Alejovigno (Talk | contribs) |
||
| Line 24: | Line 24: | ||
<h3>Constraint-based modelling</h3><br/> | <h3>Constraint-based modelling</h3><br/> | ||
| - | Constraint-based modelling use models of the cell metabolism that are derived from a metabolic network (stoicheometry models) and assume steady-state for the intracellular metabolites. These two constraints are the base of Cconstraint-based modelling: the fact that cells are subject to constraints that limit their behaviour | + | Constraint-based modelling use models of the cell metabolism that are derived from a metabolic network (stoicheometry models) and assume steady-state for the intracellular metabolites. These two constraints are the base of Cconstraint-based modelling: the fact that cells are subject to constraints that limit their behaviour [Palsson06]. In principle, if all constraints operating under |
a given set of circumstances were known, the actual state of a metabolic network | a given set of circumstances were known, the actual state of a metabolic network | ||
could be elucidated. So by imposing the known constraints, it is possible to determine which functional states can and cannot be achieved by a cell. | could be elucidated. So by imposing the known constraints, it is possible to determine which functional states can and cannot be achieved by a cell. | ||
| Line 52: | Line 52: | ||
<h3>Flux Balance Analysis</h3><br/> | <h3>Flux Balance Analysis</h3><br/> | ||
| - | <p>Flux balance analysis (FBA) is a | + | <p> |
| + | Flux balance analysis (FBA) is a methodology that uses optimisation to get predictions | ||
| + | from a constraint-based model by invoking an assumption of optimal cell behaviour. Basically, | ||
| + | one particular state among those that cells can show, accordingly to a constraint-based | ||
| + | model, is chosen based on the assumption that cells have evolved to be optimal, i.e., | ||
| + | that cells regulate its fluxes toward optimal flux states. | ||
| + | The following procedure is used to develop a flux balance analysis model: | ||
| + | </p><br/></html> | ||
| + | [[File:FBA_steps.png|600px|center|Procedure to develop a flux balance analysis model. From: PhD Thesis. F Llaneras. UPV. 2010]]<html> | ||
| + | <p> | ||
| + | |||
| + | Flux balance analysis is used to investigate hypothesis (e.g., test if a | ||
| + | reduced uptake capacity can be the cause of an unexpected cell behaviour) and to | ||
| + | evaluate a range of possibilities (e.g, find the best combination of substrates). | ||
| + | </p><br/> | ||
| + | |||
| + | <h4>Metabolic objectives and optimization</h4><br/> | ||
| + | It must be taken into account that FBA predictions, the optimal flux state, may not | ||
| + | correspond to the actual fluxes exhibit by cells. To support the assumption of optimal | ||
| + | behaviour, it must be hypothesised that: (i) cells, forced by evolutionary pressure, | ||
| + | evolved to achieve an optimal behaviour with respect to certain objective, (ii) we know | ||
| + | which this objective is, and (iii) the objective can be expressed, at least approximately, | ||
| + | in convenient mathematical terms. | ||
| + | |||
| + | Clearly, predictions of flux balance analysis are dependent on the objective function | ||
| + | being used. To date, the most commonly used objective function has been the maximisation | ||
| + | of biomass, which leaded to predictions consistent with experimental data | ||
| + | for different organisms, such as <i>Escherichia coli</i> [Edwards01]. | ||
| + | |||
| + | FBA is widely used for predicting metabolism, in particular the genome-scale metabolic network reconstructions that have been built in the past decade. These reconstructions contain all of the known metabolic reactions in an organism or plant, and the genes that encode each enzyme. In our case, FBA will calculate the flow of metabolites through our metabolic network to obtain the major production of pheromone. Must be noted that plant genome-scale models are very rare and indeed plant FBA is a very new research topic. One of the only available models is the ARAGem, a genome-scale of <i>Arabidopsis Thaliana</i></p><br/> | ||
| - | |||
| - | |||
| - | |||
<h3>References</h3> <br/> | <h3>References</h3> <br/> | ||
<p>[Llaneras08] Llaneras F, Picó J (2008). Stoichiometric Modelling of Cell Metabolism. Journal | <p>[Llaneras08] Llaneras F, Picó J (2008). Stoichiometric Modelling of Cell Metabolism. Journal | ||
of Bioscience and Bioengineering, 105:1.</p> | of Bioscience and Bioengineering, 105:1.</p> | ||
| + | <p>[Palsson06] Palsson BO (2006). Systems biology: properties of reconstructed networks. New York, USA: | ||
| + | Cambridge University Press New York.</p> | ||
| + | <p>[Edwards01] Edwards JS, Covert M, Palsson B (2002). Metabolic modelling of microbes: the | ||
| + | flux-balance approach. Environmental Microbiology, 4:133-140.</p> | ||
</div> | </div> | ||
Revision as of 21:53, 12 October 2014

Modeling Pheromone Production Overview
Pheromones production rates can be estimated using constrained-based modelling of metabolic networks.This can be useful to know about the amount of pheromone that can be produced by the synthetic plant.
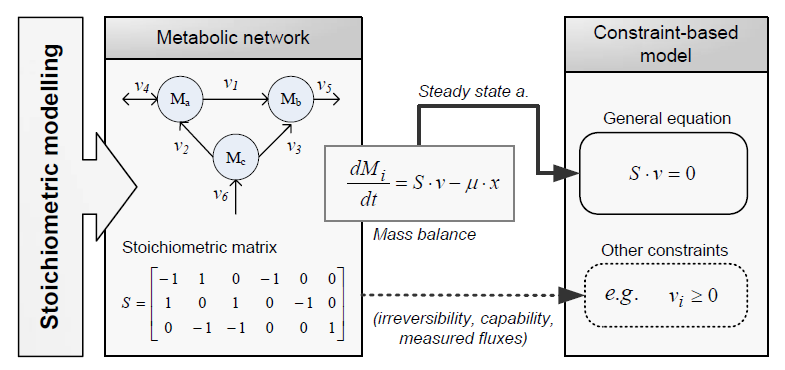
Constraint-based modelling
Constraint-based modelling use models of the cell metabolism that are derived from a metabolic network (stoicheometry models) and assume steady-state for the intracellular metabolites. These two constraints are the base of Cconstraint-based modelling: the fact that cells are subject to constraints that limit their behaviour [Palsson06]. In principle, if all constraints operating under a given set of circumstances were known, the actual state of a metabolic network could be elucidated. So by imposing the known constraints, it is possible to determine which functional states can and cannot be achieved by a cell.
Types of contraints
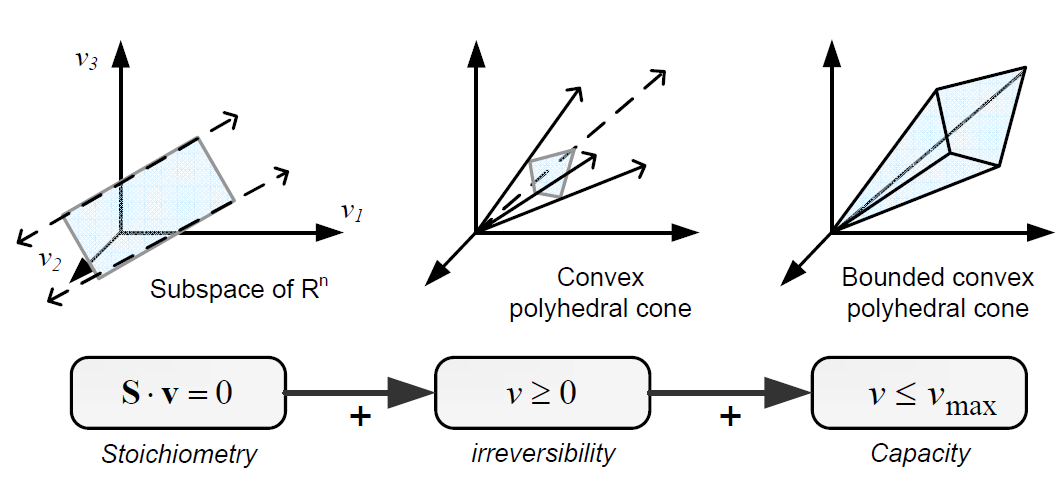
Constraints can be divided in two main types: non adjustable (invariant) and adjustable ones. The former are time-invariant restrictions of possible cell behaviour, whereas the latter depend on environmental conditions, may change through evolution, and may vary from one individual cell to another. Examples of non adjustable constraints are those imposed by thermodynamics (e.g, irreversibility of fluxes) and enzyme or transport capacities (e.g, maximum flux values). Enzyme kinetics, regulation, and experimental measurements are examples of adjustable constraints. To study the invariant properties of a network, only invariant constraints can be used, because they are those that are always satisfied (i.e, they limit the cell capabilities). If adjustable constraints are used, the elucidated cell states will be only valid under the particular set of circumstances in which these constraints operate.
Flux Balance Analysis
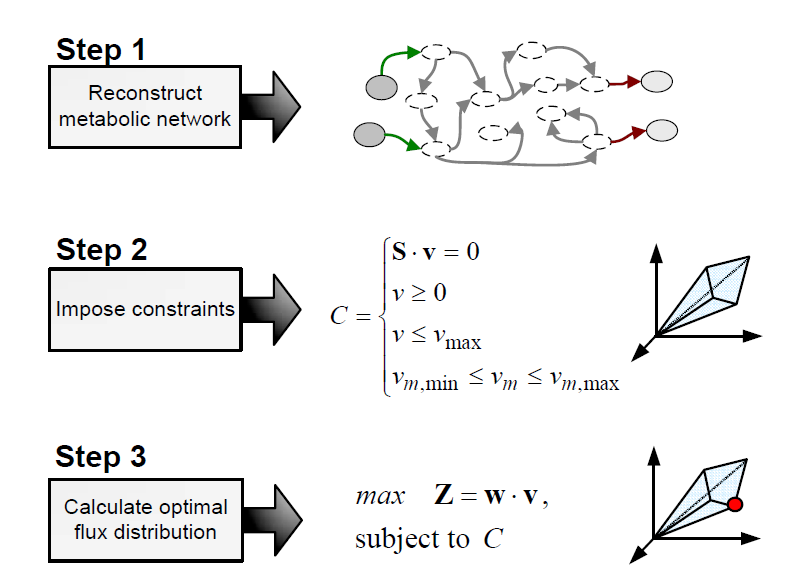
Flux balance analysis (FBA) is a methodology that uses optimisation to get predictions from a constraint-based model by invoking an assumption of optimal cell behaviour. Basically, one particular state among those that cells can show, accordingly to a constraint-based model, is chosen based on the assumption that cells have evolved to be optimal, i.e., that cells regulate its fluxes toward optimal flux states. The following procedure is used to develop a flux balance analysis model:
Flux balance analysis is used to investigate hypothesis (e.g., test if a reduced uptake capacity can be the cause of an unexpected cell behaviour) and to evaluate a range of possibilities (e.g, find the best combination of substrates).
Metabolic objectives and optimization
It must be taken into account that FBA predictions, the optimal flux state, may not correspond to the actual fluxes exhibit by cells. To support the assumption of optimal behaviour, it must be hypothesised that: (i) cells, forced by evolutionary pressure, evolved to achieve an optimal behaviour with respect to certain objective, (ii) we know which this objective is, and (iii) the objective can be expressed, at least approximately, in convenient mathematical terms. Clearly, predictions of flux balance analysis are dependent on the objective function being used. To date, the most commonly used objective function has been the maximisation of biomass, which leaded to predictions consistent with experimental data for different organisms, such as Escherichia coli [Edwards01]. FBA is widely used for predicting metabolism, in particular the genome-scale metabolic network reconstructions that have been built in the past decade. These reconstructions contain all of the known metabolic reactions in an organism or plant, and the genes that encode each enzyme. In our case, FBA will calculate the flow of metabolites through our metabolic network to obtain the major production of pheromone. Must be noted that plant genome-scale models are very rare and indeed plant FBA is a very new research topic. One of the only available models is the ARAGem, a genome-scale of Arabidopsis Thaliana
References
[Llaneras08] Llaneras F, Picó J (2008). Stoichiometric Modelling of Cell Metabolism. Journal of Bioscience and Bioengineering, 105:1.
[Palsson06] Palsson BO (2006). Systems biology: properties of reconstructed networks. New York, USA: Cambridge University Press New York.
[Edwards01] Edwards JS, Covert M, Palsson B (2002). Metabolic modelling of microbes: the flux-balance approach. Environmental Microbiology, 4:133-140.
NetLogo is an agent-based programming language and integrated modeling environment. NetLogo is free and open source software, under a GPL license.
NetLogo is an agent-based programming language and integrated modeling environment. NetLogo is free and open source software, under a GPL license.
Tab #4 content goes here!
Donec pulvinar neque sed semper lacinia. Curabitur lacinia ullamcorper nibh; quis imperdiet velit eleifend ac. Donec blandit mauris eget aliquet lacinia! Donec pulvinar massa interdum risus ornare mollis. In hac habitasse platea dictumst. Ut euismod tempus hendrerit. Morbi ut adipiscing nisi. Etiam rutrum sodales gravida! Aliquam tellus orci, iaculis vel.
 "
"