Team:SUSTC-Shenzhen/Notebook/CRISPR/mCherry-BbsI-pointmutation
From 2014.igem.org
| Line 14: | Line 14: | ||
Plasmid used: pBX-084 PB5-HS4-TRE-mCherrynuc-2A-BlaloxN-GpA-HS4-PB3 (from Wei Huang’s lab). <span style="color: red">Total length is 6843bp.</span> | Plasmid used: pBX-084 PB5-HS4-TRE-mCherrynuc-2A-BlaloxN-GpA-HS4-PB3 (from Wei Huang’s lab). <span style="color: red">Total length is 6843bp.</span> | ||
| - | Point mutation primer design: | + | Point mutation primer design:<br> |
| - | + | Forward(5’-3’): cagcccatggttttcttctgcattacggggccg 33nt<br> | |
| - | Forward(5’-3’): cagcccatggttttcttctgcattacggggccg 33nt | + | Reverse(5’-3’): cggccccgtaatgcagaagaaaaccatgggctg 33nt<br> |
| - | + | ||
| - | Reverse(5’-3’): cggccccgtaatgcagaagaaaaccatgggctg 33nt | + | |
| - | + | ||
The primer pair is intended to eliminate the BbsI site in the mCherry because we need to use BbsI to insert our gRNA into our plasmid. Designed by Agilent Technologies point-mutation primer online design tools. The primers are complement with each other. | The primer pair is intended to eliminate the BbsI site in the mCherry because we need to use BbsI to insert our gRNA into our plasmid. Designed by Agilent Technologies point-mutation primer online design tools. The primers are complement with each other. | ||
| - | |||
=Agilent's Pointmutation method= | =Agilent's Pointmutation method= | ||
| Line 351: | Line 347: | ||
Forward(5’-3’): TCGTTTAGTGAACCGTCAG 19nt<br> | Forward(5’-3’): TCGTTTAGTGAACCGTCAG 19nt<br> | ||
Reverse(5’-3’): cgaggaattcctattaTTCCTCTGCCCTCACTAG 34nt<br> | Reverse(5’-3’): cgaggaattcctattaTTCCTCTGCCCTCACTAG 34nt<br> | ||
| + | ==PCR== | ||
'''50ul, 25ul/reaction:'''<br> | '''50ul, 25ul/reaction:'''<br> | ||
''Reaction 1:'' | ''Reaction 1:'' | ||
| Line 382: | Line 379: | ||
| - | | - | ||
|} | |} | ||
| - | |||
''Reaction 2:'' | ''Reaction 2:'' | ||
{| class="table" | {| class="table" | ||
| Line 413: | Line 409: | ||
| - | | - | ||
|} | |} | ||
| - | |||
'''PCR reaction conditions:''' | '''PCR reaction conditions:''' | ||
{| class="table" | {| class="table" | ||
| Line 458: | Line 453: | ||
| 0.5ul | | 0.5ul | ||
|} | |} | ||
| - | |||
'''Adding samples''' | '''Adding samples''' | ||
{| class="table" | {| class="table" | ||
| Line 475: | Line 469: | ||
DNA marker (Takara DL5000, diluted): 30ul<br> | DNA marker (Takara DL5000, diluted): 30ul<br> | ||
| + | {{SUSTC-Image|wiki/images/d/de/SUSTC-Shenzhen-mCherry-BbsI-pointmutation-first-PCR-result.jpg}} | ||
'''Results:'''<br> | '''Results:'''<br> | ||
| Line 485: | Line 480: | ||
#The elongation time might be too long. | #The elongation time might be too long. | ||
| - | <span style="color: red">'''So I redid the 2-pari primers PCR.'''</span> | + | <span style="color: red">'''So I redid the 2-pari primers PCR with modified PCR reaction conditions.'''</span> |
| + | |||
| + | ==PCR II== | ||
| + | Reaction conditions: | ||
| + | {| class="table" | ||
| + | ! Step | ||
| + | ! Temp (℃) | ||
| + | ! Time (s) | ||
| + | |- | ||
| + | | Initial denaturation | ||
| + | | 98 | ||
| + | | 60 | ||
| + | |- | ||
| + | | rowspan="3" | 25 cycles | ||
| + | | 98 | ||
| + | | 20 | ||
| + | |- | ||
| + | | 61 | ||
| + | | 15 | ||
| + | |- | ||
| + | | 72 | ||
| + | | 15 | ||
| + | |- | ||
| + | | Final extension | ||
| + | | 72 | ||
| + | | 120 | ||
| + | |- | ||
| + | | Hold | ||
| + | | 4 | ||
| + | | ∞ | ||
| + | |} | ||
| + | |||
| + | Gel electrophoresis: | ||
| + | {{SUSTC-Image|wiki/images/c/c2/SUSTC-Shenzhen-mCherry-BbsI-pointmutation-second-PCR-result.jpg}} | ||
| + | ''Results:''<br> | ||
| + | No band is observed.<br> | ||
| + | ''Analysis:''<br> | ||
| + | #The elongation time might be too short… | ||
| + | #The annealing time for 1B might be wrong. | ||
| + | |||
| + | =2-pari primers PCR II= | ||
| + | 8.16 10:25am~11.25am<br> | ||
| + | I set a temperature gradient (45, 48, 51, 54, 57, 60℃) without shortening the elongation time. | ||
| + | ==PCR== | ||
| + | 150ul, 25ul/reaction: | ||
| + | Reaction 1: | ||
| + | Component Volume Final Concentration | ||
| + | Q5 master mix 2X 75ul 1X | ||
| + | pBX084 diluted (25.14ng/ul) 4ul 100.56ng | ||
| + | mCherry Primer F (50uM) 2ul 333.3nM | ||
| + | Pointmutation Primer R (50uM) 2ul 333.3nM | ||
| + | ddH20 67ul - | ||
| + | Total 150ul - | ||
| + | Reaction 2: | ||
| + | Component Volume Final Concentration | ||
| + | Q5 master mix 2X 75ul 1X | ||
| + | pBX084 diluted (25.14ng/ul) 4ul 100.56ng | ||
| + | mCherry Primer R (50uM) 2ul 333.3nM | ||
| + | Pointmutation Primer F (50uM) 2ul 333.3nM | ||
| + | ddH20 67ul - | ||
| + | Total 150ul - | ||
PCR reaction conditions: | PCR reaction conditions: | ||
Step Temp (℃) Time (s) | Step Temp (℃) Time (s) | ||
Initial denaturation 98 60 | Initial denaturation 98 60 | ||
25 cycles 98 20 | 25 cycles 98 20 | ||
| - | + | 45, 48, 51, 54, 57, 60 15 | |
| - | 72 | + | 72 20 |
| - | Final extension 72 | + | Final extension 72 300 |
Hold 4 ∞ | Hold 4 ∞ | ||
| + | |||
| + | 11:50am ~12:25pm Gel electrophoresis | ||
| + | |||
| + | Unfortunately, only in A2 did I observe the band. I doubt the annealing temperature was not high enough because strong primer dimers forms in 1B group. So I redid the 2-pair primers PCR again with even higher temperature gradient (62,63,64,65,66,67℃). | ||
Gel electrophoresis: | Gel electrophoresis: | ||
Results: | Results: | ||
| - | + | Only vague bands were observed in A2. Still no distinct bands observed in 1B. | |
Analysis: | Analysis: | ||
| - | 1. The | + | 1. The reverse primer of pointmutation might be very easy to form dimer with the forward primer of mCherry, which led to the failure of PCR. |
| - | 2. The | + | 2. The compatibility of the two separate primers might be very bad. |
| + | I decided to try another methods. | ||
{{SUSTC-Shenzhen/main-content-end}} | {{SUSTC-Shenzhen/main-content-end}} | ||
{{SUSTC-Shenzhen/wiki-footer}} | {{SUSTC-Shenzhen/wiki-footer}} | ||
{{SUSTC-Shenzhen/themeJs}} | {{SUSTC-Shenzhen/themeJs}} | ||
Revision as of 14:52, 12 October 2014
mCherry BbsI point mutation
one step for constructing a universal usable gRNA-insertion plasmid
Contents |
By Yicong Tao
Plasmid used: pBX-084 PB5-HS4-TRE-mCherrynuc-2A-BlaloxN-GpA-HS4-PB3 (from Wei Huang’s lab). Total length is 6843bp.
Point mutation primer design:
Forward(5’-3’): cagcccatggttttcttctgcattacggggccg 33nt
Reverse(5’-3’): cggccccgtaatgcagaagaaaaccatgggctg 33nt
The primer pair is intended to eliminate the BbsI site in the mCherry because we need to use BbsI to insert our gRNA into our plasmid. Designed by Agilent Technologies point-mutation primer online design tools. The primers are complement with each other.
Agilent's Pointmutation method
8.13~8.14
PCR reaction
(12:06~14:XX 13th Aug, about 2h)
300ul, 50ul/reaction:
| Component | Volume | Final Concentration |
|---|---|---|
| Q5 master mix 2X | 150ul | 1X |
| DNA template (251.4ng/ul) | 1.5ul | 1.257ng/ul |
| Primer F (50uM) | 3ul | 500nM |
| Primer R (50uM) | 3ul | 500nM |
| ddH20 | 142.5ul | - |
| Total | 300ul | - |
PCR reaction conditions:
| Step | Temp (℃) | Time (s) |
|---|---|---|
| Initial denaturation | 98 | 60 |
| 18 cycles | 98 | 10 |
| 58, 60, 61, 62, 63, 65 | 15 | |
| 72 | 360 | |
| Final extension | 72 | 120 |
| Hold | 4 | ∞ |
DpnI digestion
(13 Aug 15:05~19:06)
Add 1ul DpnI directly to the amplification system
Digest 4h
No termination
Stored at -20℃
Nanodrop test after digestion
| Anneal Temp (℃) | 58 | 60 | 61 | 62 |
|---|---|---|---|---|
| Conc (ng/ul) | 467.4 | 465.9 | 470.4 | 457.9 |
| 260/280 | 1.8 | 1.8 | 1.8 | 1.81 |
| 260/230 | 0.73 | 0.73 | 0.74 | 0.73 |
Bacteria transformation
(2014.8.13 22:08~2014.8.14 14:50)
Procedures:
- Immediately, place the tubes on ice, allow cell to thaw on ice, 10 min
- Check the cell to see if they have thawed, gently flick the cells 1-2 times to evenly resuspend the cells.
- Divide the competent cells into 6 tubes,50μL each tube, Mark them with 58, 60, 61, 62, 63, 65. Keep the bacteria in ice during the procedure.
- Add 5μL plasmid to each tube, shaking slightly
- Incubate on ice for 30min
- Heat shock, 42°C, 90s.
- Keep on ice, 2 min
- Add 200μl SOC medium to each tube, incubate 37°C, 200rpm, 45min
- Centrifuge, 4000rmp, 5min, RT. Discard most of the medium and reserve 50μL. Resuspend.
- Add 50μL bacteria to amp LB agar plate, distribute.
- Incubate the plates at 37°C, 16 hours.
Results:
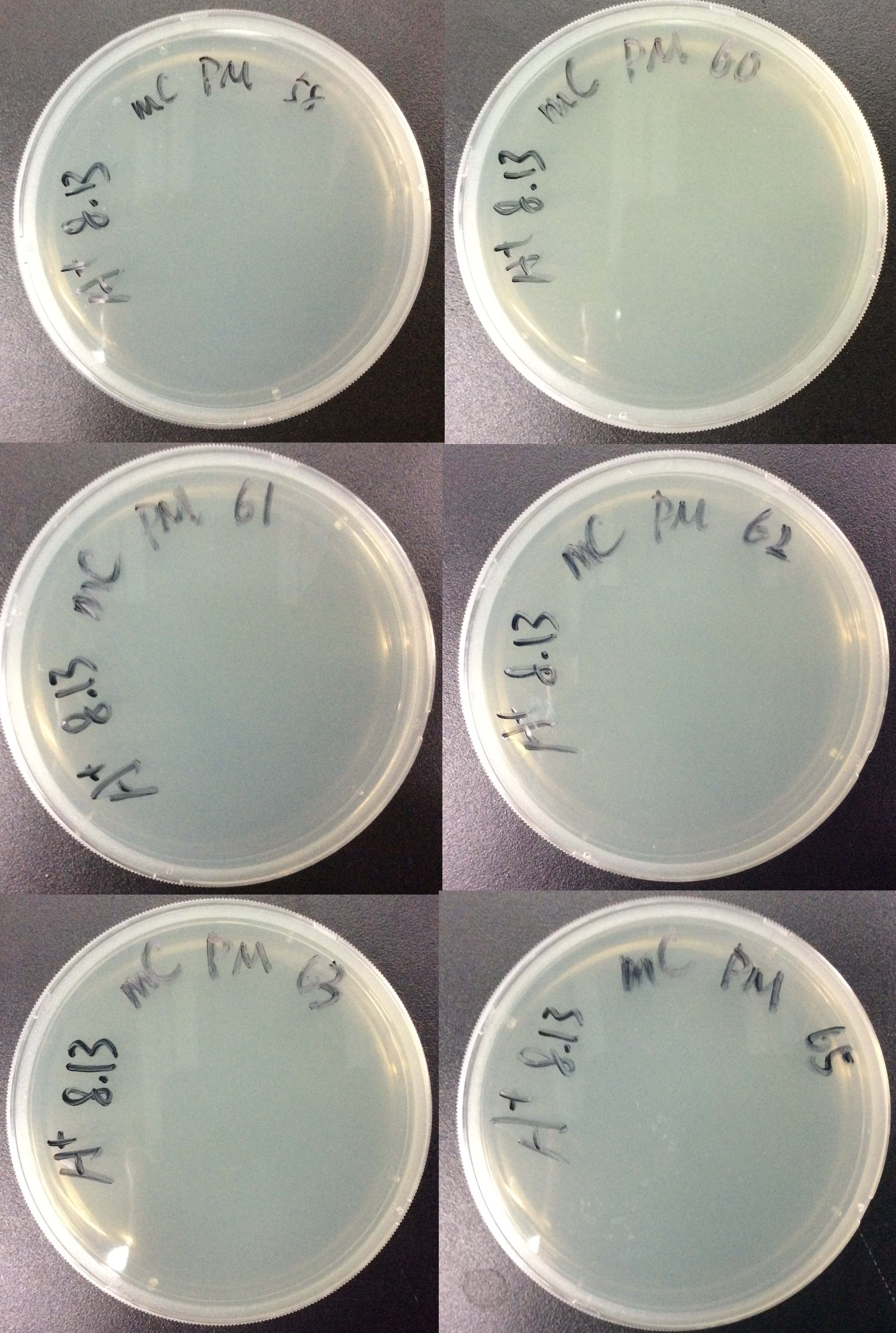
For all 6 groups, no colony forms.
Gel electrophoresis
Making the agarose gel (1%)
| Component | Volume |
|---|---|
| TAE | 25ml |
| Agarose | 0.25g |
| Gene Green | 0.5ul |
Adding samples
| Component | Volume |
|---|---|
| PCR product | 1ul |
| 6X loading dye | 2ul |
| TAE | 9ul |
| Total | 12ul |
DNA marker (Takara DL5000, diluted): 10ul
Results:
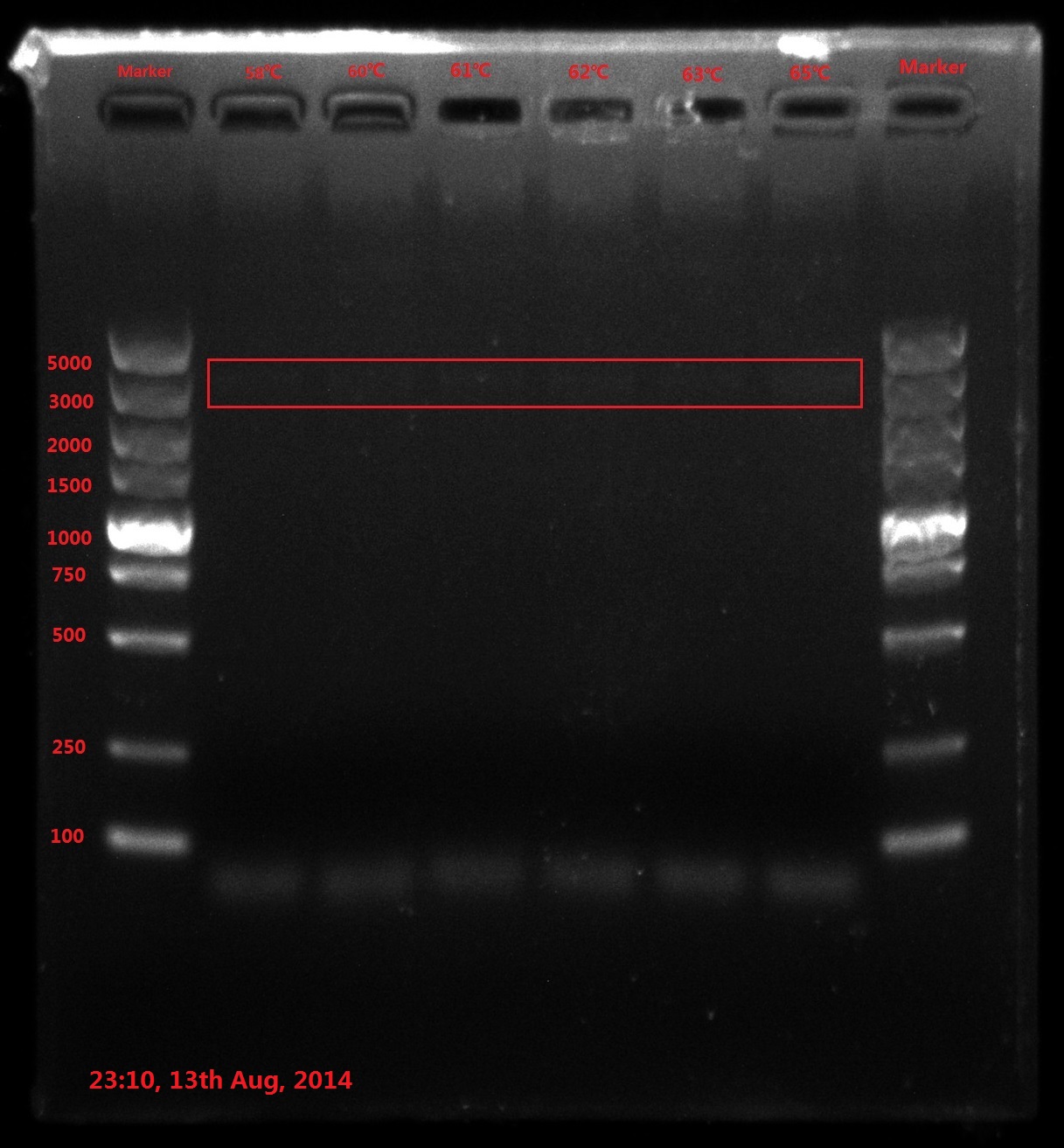
The result of electrophoresis is not clear, so I redid the gel electrophoresis with the same sample.
No wanted band (6843bp) is observed.
Discussion:
- PCR failed, so improving PCR conditions is the first choice. Try extending PCR cycles, denaturation time, annealing time and final extension time.
- Do reaction termination after DpnI enzyme digestion (80℃ 20min) next time because reactive enzyme may binding to the DNA substrate and interfere the transformation efficiency.
- The concentration of template DNA is also very essential, so using 2-step concentration ladder next time.
Agilent's Pointmutation method II
8.15
PCR
(23:35 14th Aug-08:30 15th Aug), about 2h. Next morning, check PCR machine was not at 4℃ hold conditions, and the heating stand was not cold
200ul, 50ul/reaction:
| Component | Volume | Final Concentration |
|---|---|---|
| Q5 master mix 2X | 100ul | 1X |
| DNA template (251.4ng/ul) | 1.0ul (A,B), 2.0ul (C,D) | 1.257ng/ul (A,B), 2.514ng/ul (C,D) |
| Primer F (50uM) | 2ul | 500nM |
| Primer R (50uM) | 2ul | 500nM |
| ddH20 | 95ul | - |
| Total | 200ul | - |
PCR reaction conditions:
| Step | Temp (℃) | Time (s) |
|---|---|---|
| Initial denaturation | 98 | 60 |
| 25 cycles | 98 | 20 |
| 63 | 30 | |
| 72 | 420 | |
| Final extension | 72 | 600 |
| Hold | 4 | ∞ |
Nanodrop test
| Anneal Temp (℃) | A | B | C | D |
|---|---|---|---|---|
| Conc (ng/ul) | 474.2 | 492.0 | 466.7 | 445.9 |
| 260/280 | 1.82 | 1.81 | 1.81 | 1.81 |
| 260/230 | 0.72 | 0.73 | 0.72 | 0.70 |
Gel electrophoresis
Making the agarose gel (1%)
| Component | Volume |
|---|---|
| TAE | 25ml |
| Agarose | 0.25g |
| Gene Green | 0.5ul |
Adding samples
| Component | Volume |
|---|---|
| PCR product | 5ul |
| 6X loading dye | 2ul |
| TAE | 5ul |
| Total | 12ul |
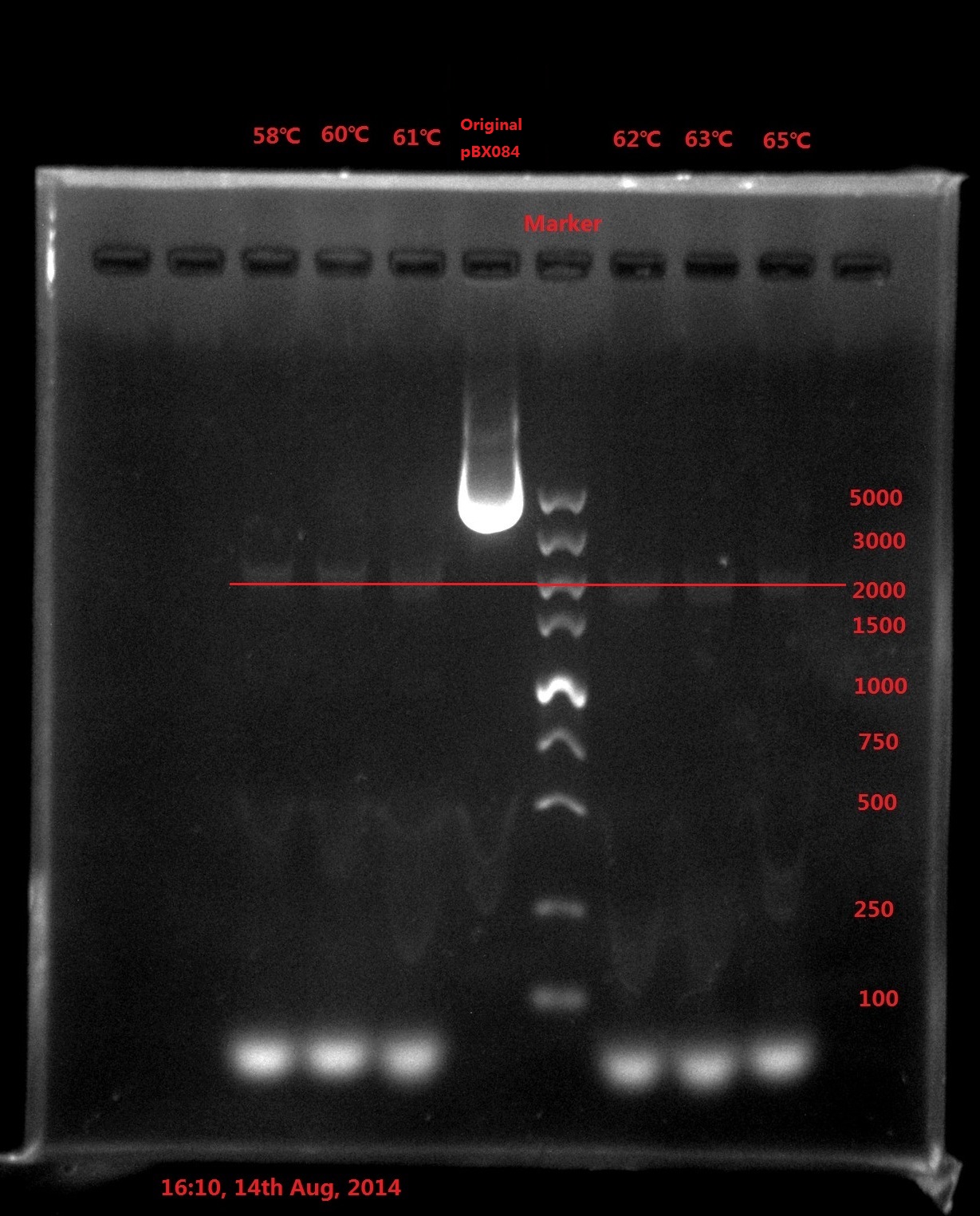
DNA marker (Takara DL5000, diluted): 10ul
Results:
Analysis:
The band was in 2000bp location, which is far away from the original plasmid length (6800bp), which meant PCR didn’t got complete plasmid.
The possible reason might be:
- The primer forms dimers which strong influence the efficiency of PCR (The smallest band is very light).
- DNA is a double helix, which means the plasmid double strand can also anneal with each other. The elongation time was very long, which left enough time for the plasmid double strand to anneal with each other and prohibit the PCR process.
Solution:
- Redesign the primer so that the primer pairs are not totally complementary with each other. Put the point mutation point near the 5’ end.
- Use other PCR methods.
So I decided to change the PCR method.
2-pair primers PCR
I had two pairs of primers: one is for mCherry (called A), another is for point mutation (called B). I then used the forward primer of A to pair with the reverse primer of B, so did the forward primer of B with the reverse primer of A. Thus, I would get 2 short segments which had a small complementary region. Then, I added the two segments together and let them do PCR using each other as the template and primer in order to get the final whole length PCR product.
mCherry primer design:
Forward(5’-3’): TCGTTTAGTGAACCGTCAG 19nt
Reverse(5’-3’): cgaggaattcctattaTTCCTCTGCCCTCACTAG 34nt
PCR
50ul, 25ul/reaction:
Reaction 1:
| Component | Volume | Final Concentration |
|---|---|---|
| Q5 master mix 2X | 25ul | 1X |
| pBX084 diluted (25.14ng/ul) | 2.0ul | 1.257ng/ul |
| mCherry Primer F (50uM) | 0.5ul | 500nM |
| Pointmutation Primer R (50uM) | 0.5ul | 500nM |
| ddH20 | 22ul | - |
| Total | 50ul | - |
Reaction 2:
| Component | Volume | Final Concentration |
|---|---|---|
| Q5 master mix 2X | 25ul | 1X |
| pBX084 diluted (25.14ng/ul) | 2.0ul | 1.257ng/ul |
| mCherry Primer R (50uM) | 0.5ul | 500nM |
| Pointmutation Primer F (50uM) | 0.5ul | 500nM |
| ddH20 | 22ul | - |
| Total | 50ul | - |
PCR reaction conditions:
| Step | Temp (℃) | Time (s) |
|---|---|---|
| Initial denaturation | 98 | 60 |
| 25 cycles | 98 | 20 |
| 61 | 15 | |
| 72 | 20 | |
| Final extension | 72 | 300 |
| Hold | 4 | ∞ |
Gel electrophoresis
Making the agarose gel (1%)
| Component | Volume |
|---|---|
| TAE | 25ml |
| Agarose | 0.25g |
| Gene Green | 0.5ul |
Adding samples
| Component | Volume |
|---|---|
| PCR product | 10ul |
| 6X loading dye | 2ul |
| Total | 12ul |
DNA marker (Takara DL5000, diluted): 30ul

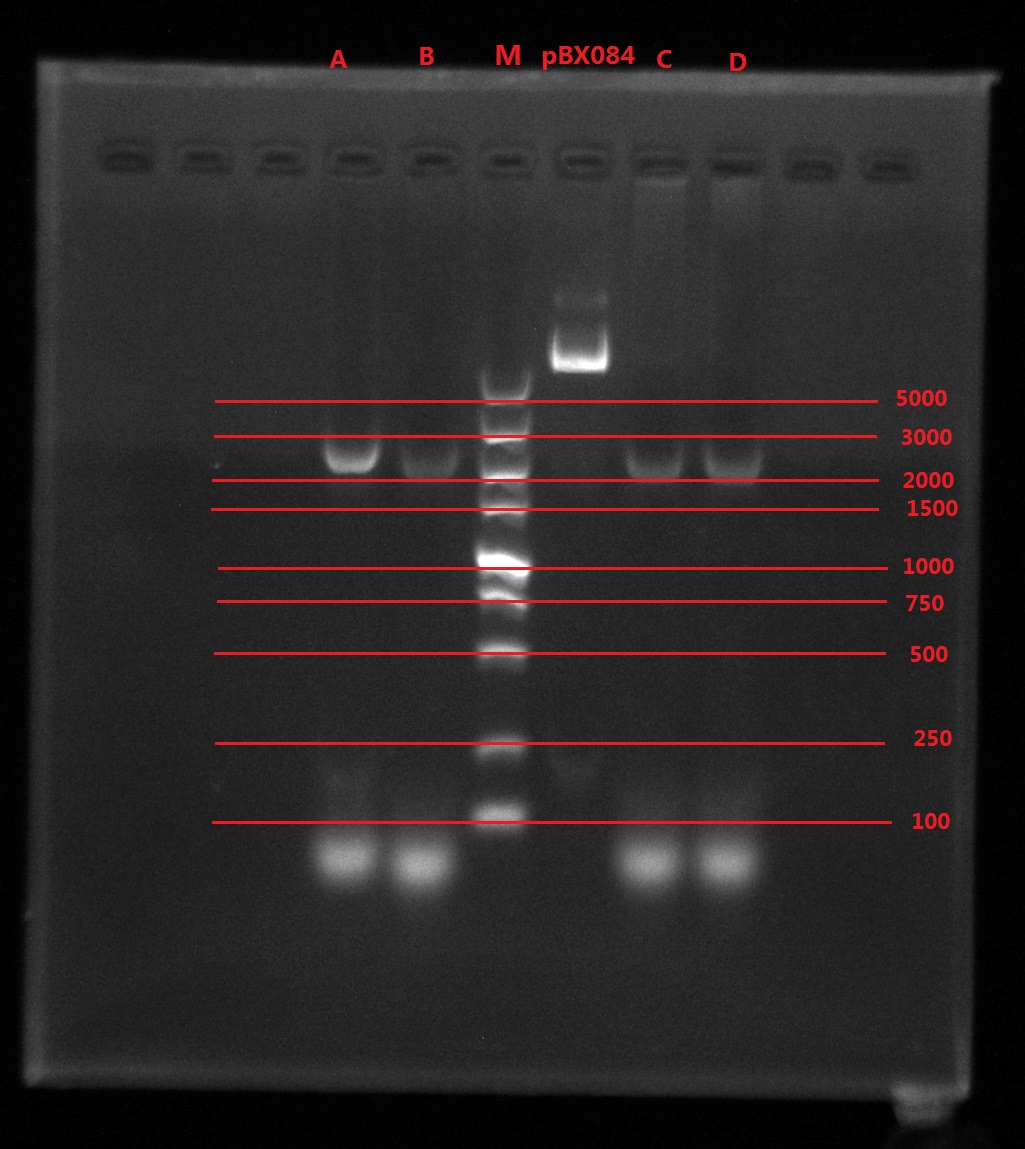
Results:
A2: reaction 1 (A: mCherry primer F; 2: Pointmutation Primer R)
1B: reaction 2 (B: mCherry primer R; 1: Pointmutation Primer F)
Analysis:
The predicting results is 448bp band in A2 and 372bp band in 1B (almost equally separate the mCherry). But the band in A2 was in 800bp and no band is observed in 1B. The possible reasons might be:
- I added wrong primer pairs: two mCherry primers for A2 and two pointmutation primers for 1B. So that in A2 I got mCherry and in 1B I got primer dimer.
- The elongation time might be too long.
So I redid the 2-pari primers PCR with modified PCR reaction conditions.
PCR II
Reaction conditions:
| Step | Temp (℃) | Time (s) |
|---|---|---|
| Initial denaturation | 98 | 60 |
| 25 cycles | 98 | 20 |
| 61 | 15 | |
| 72 | 15 | |
| Final extension | 72 | 120 |
| Hold | 4 | ∞ |
Gel electrophoresis:
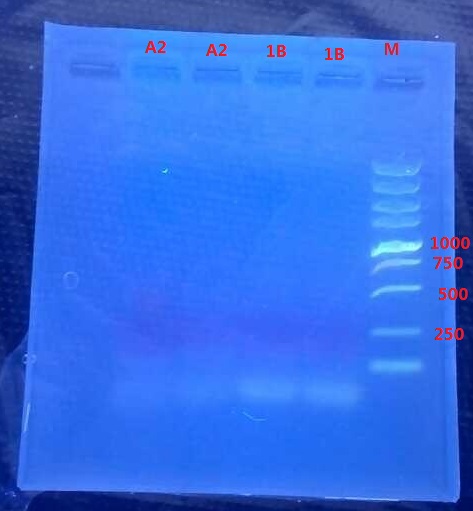 Results:
Results:
No band is observed.
Analysis:
- The elongation time might be too short…
- The annealing time for 1B might be wrong.
2-pari primers PCR II
8.16 10:25am~11.25am
I set a temperature gradient (45, 48, 51, 54, 57, 60℃) without shortening the elongation time.
PCR
150ul, 25ul/reaction: Reaction 1: Component Volume Final Concentration Q5 master mix 2X 75ul 1X pBX084 diluted (25.14ng/ul) 4ul 100.56ng mCherry Primer F (50uM) 2ul 333.3nM Pointmutation Primer R (50uM) 2ul 333.3nM ddH20 67ul - Total 150ul - Reaction 2: Component Volume Final Concentration Q5 master mix 2X 75ul 1X pBX084 diluted (25.14ng/ul) 4ul 100.56ng mCherry Primer R (50uM) 2ul 333.3nM Pointmutation Primer F (50uM) 2ul 333.3nM ddH20 67ul - Total 150ul - PCR reaction conditions: Step Temp (℃) Time (s) Initial denaturation 98 60 25 cycles 98 20 45, 48, 51, 54, 57, 60 15 72 20 Final extension 72 300 Hold 4 ∞
11:50am ~12:25pm Gel electrophoresis
Unfortunately, only in A2 did I observe the band. I doubt the annealing temperature was not high enough because strong primer dimers forms in 1B group. So I redid the 2-pair primers PCR again with even higher temperature gradient (62,63,64,65,66,67℃).
Gel electrophoresis:
Results: Only vague bands were observed in A2. Still no distinct bands observed in 1B. Analysis: 1. The reverse primer of pointmutation might be very easy to form dimer with the forward primer of mCherry, which led to the failure of PCR. 2. The compatibility of the two separate primers might be very bad.
I decided to try another methods.
 "
"