Team:SUSTC-Shenzhen/Notebook/CRISPR/mCherry-BbsI-pointmutation
From 2014.igem.org
| Line 149: | Line 149: | ||
Results: | Results: | ||
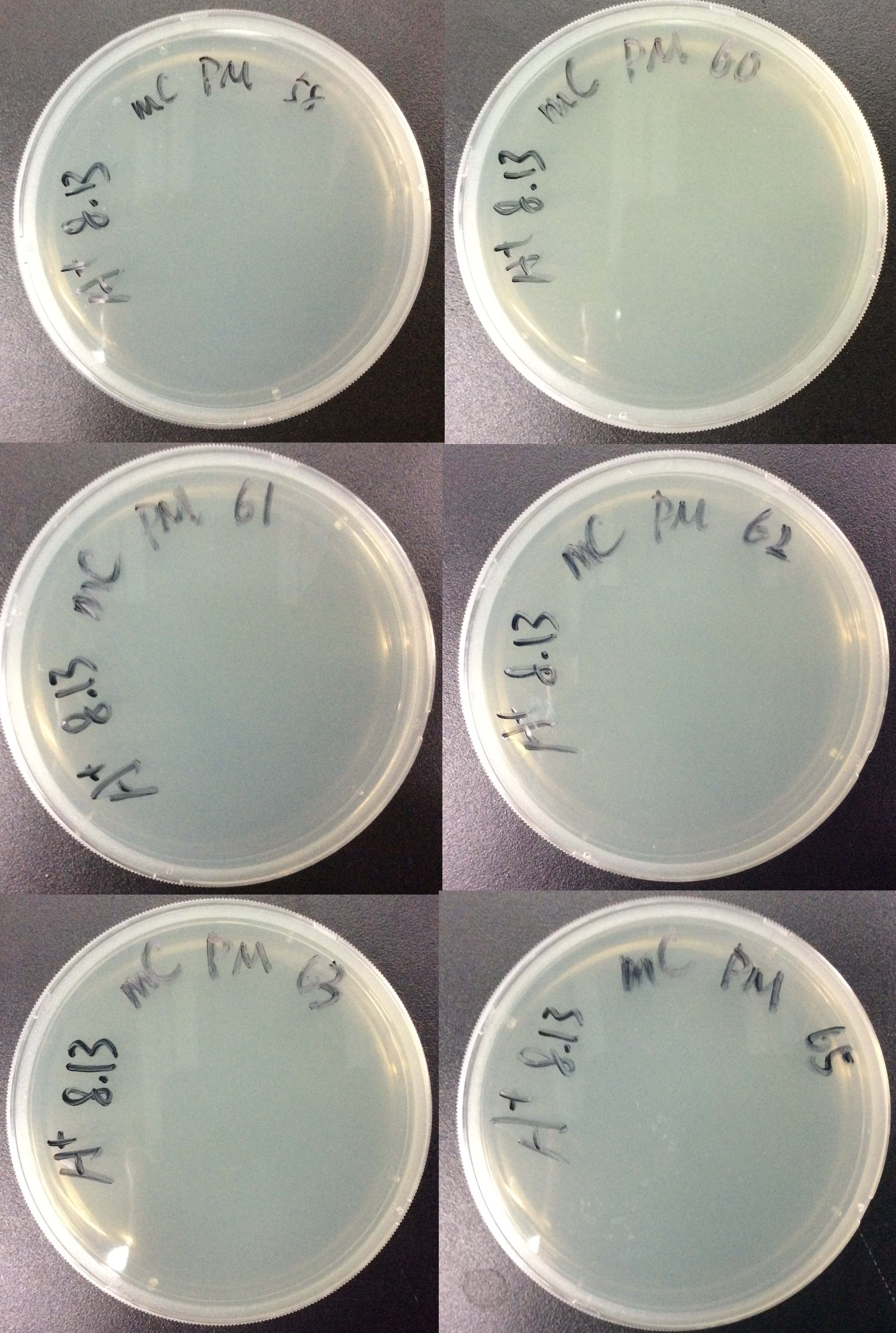
| - | {{SUSTC-Image|wiki/images/ | + | {{SUSTC-Image|wiki/images/e/e1/SUSTC-Shenzhen-mCherry-BbsI-pointmutation-colony.jpg|Bacteria transformation results}} |
| - | + | ||
| - | + | ||
For all 6 groups, no colony forms. | For all 6 groups, no colony forms. | ||
Revision as of 09:49, 12 October 2014
mCherry BbsI point mutation
one step for constructing a universal usable gRNA-insertion plasmid
Contents |
By Yicong Tao
Plasmid used: pBX-084 PB5-HS4-TRE-mCherrynuc-2A-BlaloxN-GpA-HS4-PB3 (from Wei Huang’s lab). Total length is 6843bp.
Point mutation primer design:
Forward(5’-3’): cagcccatggttttcttctgcattacggggccg 33nt
Reverse(5’-3’): cggccccgtaatgcagaagaaaaccatgggctg 33nt
The primer pair is intended to eliminate the BbsI site in the mCherry because we need to use BbsI to insert our gRNA into our plasmid. Designed by Agilent Technologies point-mutation primer online design tools. The primers are complement with each other.
8.13~8.14 Agilent's Pointmutation method
PCR reaction
(12:06~14:XX 13th Aug, about 2h)
300ul, 50ul/reaction:
| Component | Volume | Final Concentration |
|---|---|---|
| Q5 master mix 2X | 150ul | 1X |
| DNA template (251.4ng/ul) | 1.5ul | 1.257ng/ul |
| Primer F (50uM) | 3ul | 500nM |
| Primer R (50uM) | 3ul | 500nM |
| ddH20 | 142.5ul | - |
| Total | 300ul | - |
PCR reaction conditions:
| Step | Temp (℃) | Time (s) |
|---|---|---|
| Initial denaturation | 98 | 60 |
| 18 cycles | 98 | 10 |
| 58, 60, 61, 62, 63, 65 | 15 | |
| 72 | 360 | |
| Final extension | 72 | 120 |
| Hold | 4 | ∞ |
DpnI digestion
(13 Aug 15:05~19:06)
Add 1ul DpnI directly to the amplification system
Digest 4h
No termination
Stored at -20℃
Nanodrop test after digestion
| Anneal Temp (℃) | 58 | 60 | 61 | 62 |
|---|---|---|---|---|
| Conc (ng/ul) | 467.4 | 465.9 | 470.4 | 457.9 |
| 260/280 | 1.8 | 1.8 | 1.8 | 1.81 |
| 260/230 | 0.73 | 0.73 | 0.74 | 0.73 |
Bacteria transformation
(2014.8.13 22:08~2014.8.14 14:50)
Procedures:
- Immediately, place the tubes on ice, allow cell to thaw on ice, 10 min
- Check the cell to see if they have thawed, gently flick the cells 1-2 times to evenly resuspend the cells.
- Divide the competent cells into 6 tubes,50μL each tube, Mark them with 58, 60, 61, 62, 63, 65. Keep the bacteria in ice during the procedure.
- Add 5μL plasmid to each tube, shaking slightly
- Incubate on ice for 30min
- Heat shock, 42°C, 90s.
- Keep on ice, 2 min
- Add 200μl SOC medium to each tube, incubate 37°C, 200rpm, 45min
- Centrifuge, 4000rmp, 5min, RT. Discard most of the medium and reserve 50μL. Resuspend.
- Add 50μL bacteria to amp LB agar plate, distribute.
- Incubate the plates at 37°C, 16 hours.
Results:
For all 6 groups, no colony forms.
Gel electrophoresis
Making the agarose gel (1%)
| Component | Volume |
|---|---|
| TAE | 25ml |
| Agarose | 0.25g |
| Gene Green | 0.5ul |
Adding samples
| Component | Volume |
|---|---|
| PCR product | 1ul |
| 6X loading dye | 2ul |
| TAE | 9ul |
| Total | 12ul |
DNA marker (Takara DL5000, diluted): 10ul
Results:
The result of electrophoresis is not clear, so I redid the gel electrophoresis with the same sample.
No wanted band (6843bp) is observed.
Discussion: 1. PCR failed, so improving PCR conditions is the first choice. Try extending PCR cycles, denaturation time, annealing time and final extension time. 2. Do reaction termination after DpnI enzyme digestion (80℃ 20min) next time because reactive enzyme may binding to the DNA substrate and interfere the transformation efficiency. 3. The concentration of template DNA is also very essential, so using 2-step concentration ladder next time.
 "
"