lab note of circuit in the control part
part/gene transformation and test
circuit contruction
part function test
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
lab note of circuit in the communication part
part/gene transformation and test
circuit contruction
part function test
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
lab note of circuit in the cohesion part
part/gene transformation and test
circuit contruction
part function test
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
lab note of circuit in the completion part
Parts from iGEM took kit
Out line
For each part to be used in testing circuit:
BBa_J23100 , BBa_E1010, BBa_B0015
![]()
circuit 圖
After transforming these plasmids into our competent cell, DH5a, we would take 3 steps to examine if we got the right product:
- Colony PCR and electrophoresis: Directly picking up colonies on designated antibiotic plate and do colony PCR with iGEM provided primers (VF2&VR), and then do electrophoresis to check the length of plasmid to confirm if the kit had been correctly transformed into E. coli.
- Plasmid PCR and electrophoresis: Colony PCR may not end up copying the right place, for there is extra DNA in the cell that is not the one of our interest. Therefore, we also check the length of the plasmid we which we transformed.
- Digestion and electrophoresis: After checking the length, we use EcoR1/Spe1 and Xba1/Pst1, and electrophoresis, to see if our constructed plasmid can be successfully digested.
Process
We encounter several difficulties during the process:
- In the transformation step, we couldn’t get transformation with the right PCR band at first. Even thought they do grow on the antibiotic plate, electrophoresis result doesn’t show any band. Therefore, we tried different ways trying to solve these problems.
First, we thought it was PCR problem, therefore, we tried colony PCR. However, it came out that it wasn’t the case.So we improve our transformation protocol. That is, after 1.5 hour of recovery in 37C incubator in 200ml LB, we centrifuge in 3.4 rpm for 20 seconds, and discard 150ml supernatant.
At the end, we result in putting concentrated transformed E.coli into our antibiotic plate. This way, we sequentially transformed several kits successfully.
- In the checking step, we’ve have contaminated electrophoresis result after colony PCR. That is, we got band in the negative well. To find out the ingredient causing contamination, we do several negative control changing different packing of ingredients. And find the contamination of our 10X DreamTaq Buffer.
Result
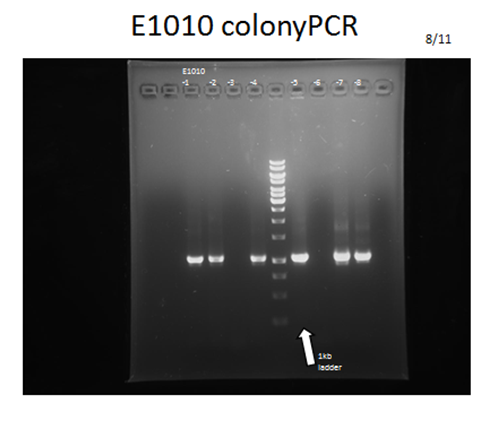
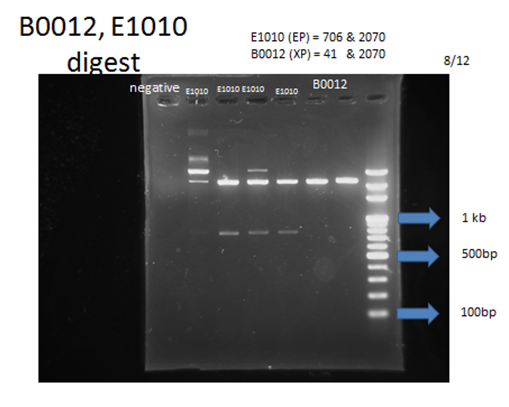
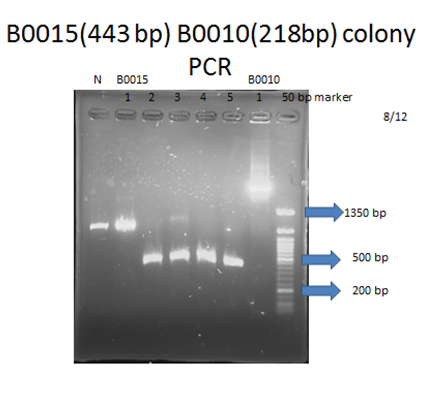
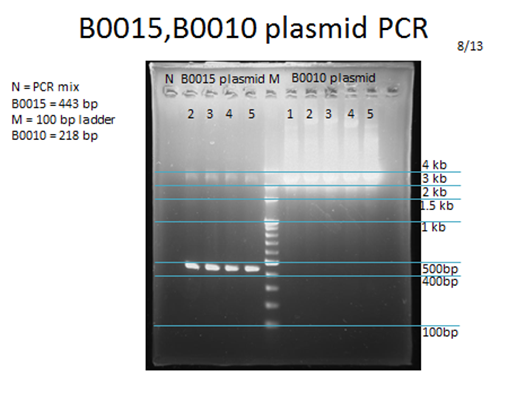
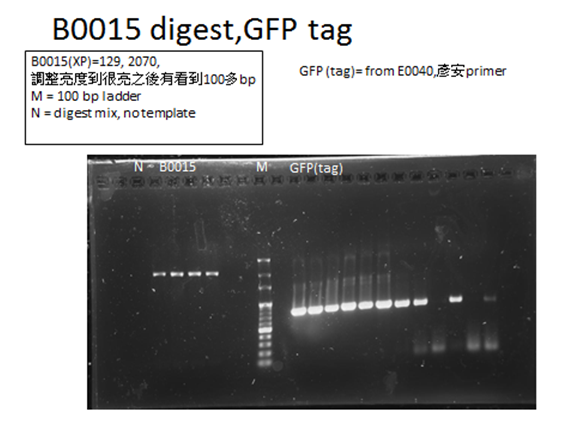
Parts that needed extra PCR
Outline
Two parts needed extra primer in order to get them:
yebF, RFP, and M102 orf19
![]()
circuit 圖
And for each part, we do two kinds of PCR , one using Dream Tag, only to check the length to ensure we got our part. And then we used KOD enzyme, together with magnesium cation, to check the length, furthermore, do gel extraction to get the part. That’s because KOD enzyme leads a more accurate PCR.
- For yebF, we designed primers to get the sequence we want yebF (its own ribosomal binding site included) from Escherichia coli MG1655.
Front Primer : tttctagagGGAGAAAAACATGAAAAAAAGAGGGGCGTT(with Xbal cutting site)
Reverse Primer : attctgcagcggccgctactagtACGCCGCTGATATTCCGCCA(with Spel and Pst1 cutting site)
The PCR product should be 389bp.
- Because the need of recombinant protein, the RFP downstream of yebF need to be specially designed in order to leave scar with multiple of 3. E1010 was used as template, and primers with specially designed cutting site is made:
Front Primer: tttctagaGCTTCCTCCTCCGAAGACGTTATC
And normal iGEM reverse primer, VR
Process
While doing RFP PCR, we got extra band.
By adjusting PCR program, we finally got correct band.
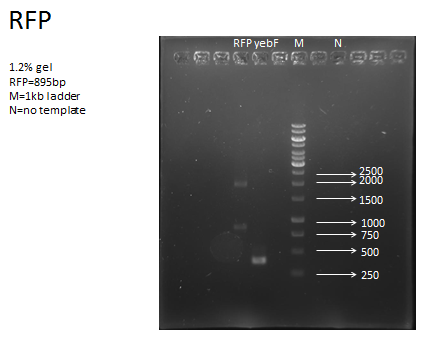
Result



Construction of the circuit
Outline
Circuits needed to be constructed
J23119-(RBS)yebF-RFP-B0015
J23119_B0034_RFP-B0015
J23119-yebF-B0015
J23119-yebF-orf19-B0015
By repeatedly using back insert, we tried to achieve the final construction.We planned to constructed our first circuit by following steps:
- J23119(SP)+yebF(XP)
- J23119+yebF(SP)+RFP(XP)
- J23119+yebF+RFP(SP)+B0015(XP)
Process
During circuit construction, we encountered several problems:
First, we the enzyme that we own are from different companies. Our digestion enzyme Pst1 is from Fast Digestion, requiring FD buffer, while the Spel enzyme is from NEB, requiring the addition of NEB buffer 4 and BSA, or smart cut buffer.
Also, because that one of the part result from cutting is so small that it’s invisible in electrophoresis, we can only check if our product had been successfully cut by transformation. We also did electrophoresis, though, to check if the band would be good enough to do extraction. Finally, by doing checking transformation product after ligation, we can fully sure that we had the right digestion cut.
Our first ligation result shows that using FD buffer is absolutely impossible if we want to digest with Spel and Pst1. As shown on the graph, it might just stick back to the original plasmid.

lab note of function test in phage
part/gene transformation and test
circuit contruction
part function test
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
under construction
experiment1 date: 0707~0712
under construction
under construction
 "
"






























