Team:Aachen/Project/Model
From 2014.igem.org
(→Modeling) |
(→Modeling) |
||
| Line 14: | Line 14: | ||
</center> | </center> | ||
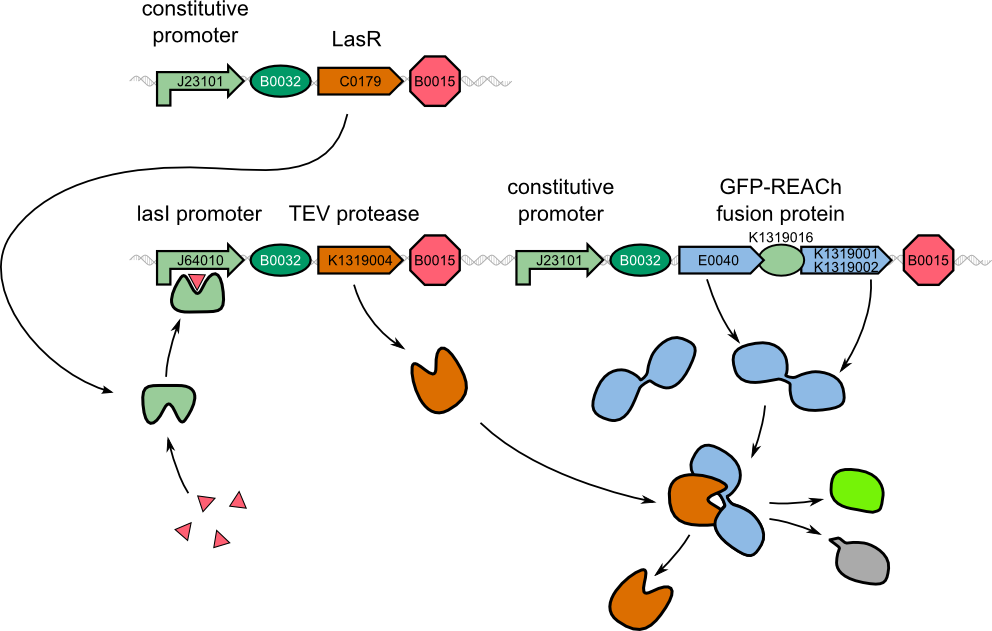
| - | For our two-dimensional biosensor, we thought of different methods to generate a faster and stronger | + | For our two-dimensional biosensor, we thought of different methods to generate a faster and stronger fluorescence response from weak promoters. In our molecular approach (left) to detect ''P. aeruginosa'', a fusion protein of GFP and the dark quencher is cleaved by the very specific TEV protease which is introduced behind the weak quorum sensing promoter. |
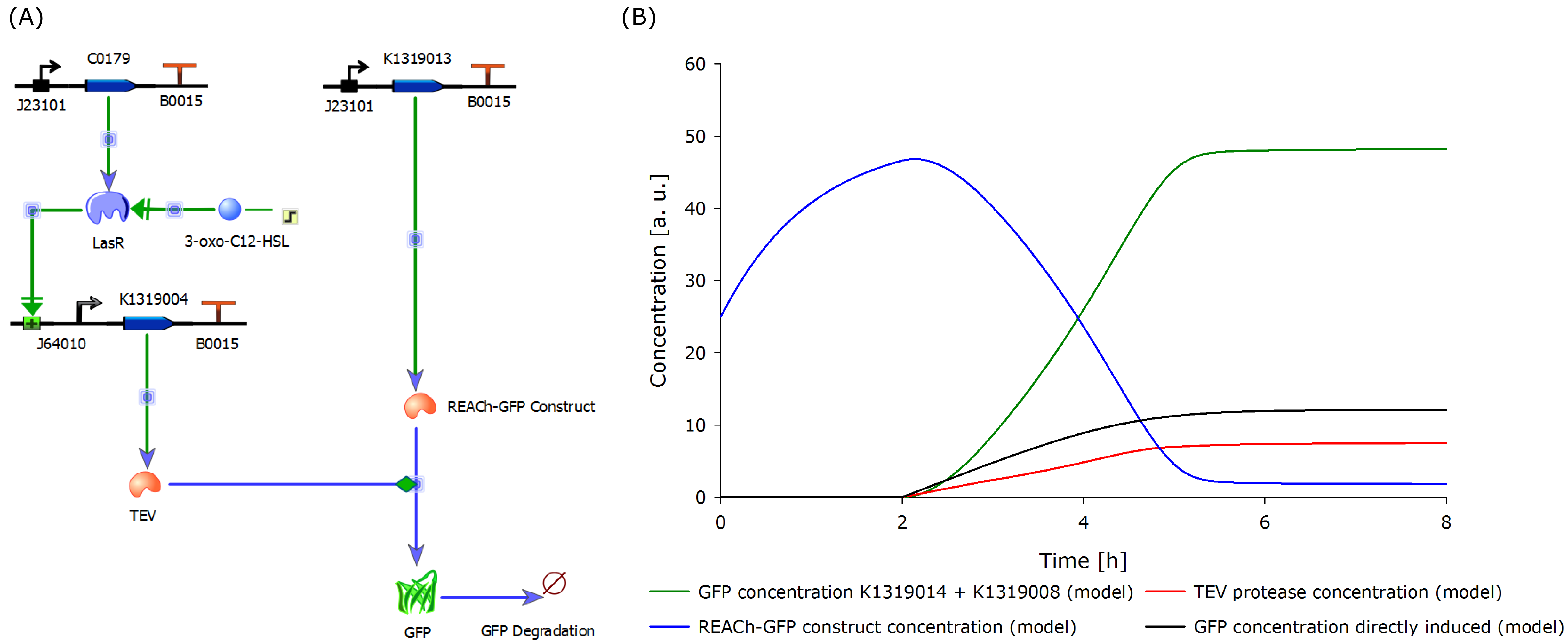
To validate our hypothesis, we developed a model of our molecular approach using the CAD tool TinkerCell, see figure below (Chandran, Bergmann and Sauro, 2009). | To validate our hypothesis, we developed a model of our molecular approach using the CAD tool TinkerCell, see figure below (Chandran, Bergmann and Sauro, 2009). | ||
| Line 35: | Line 35: | ||
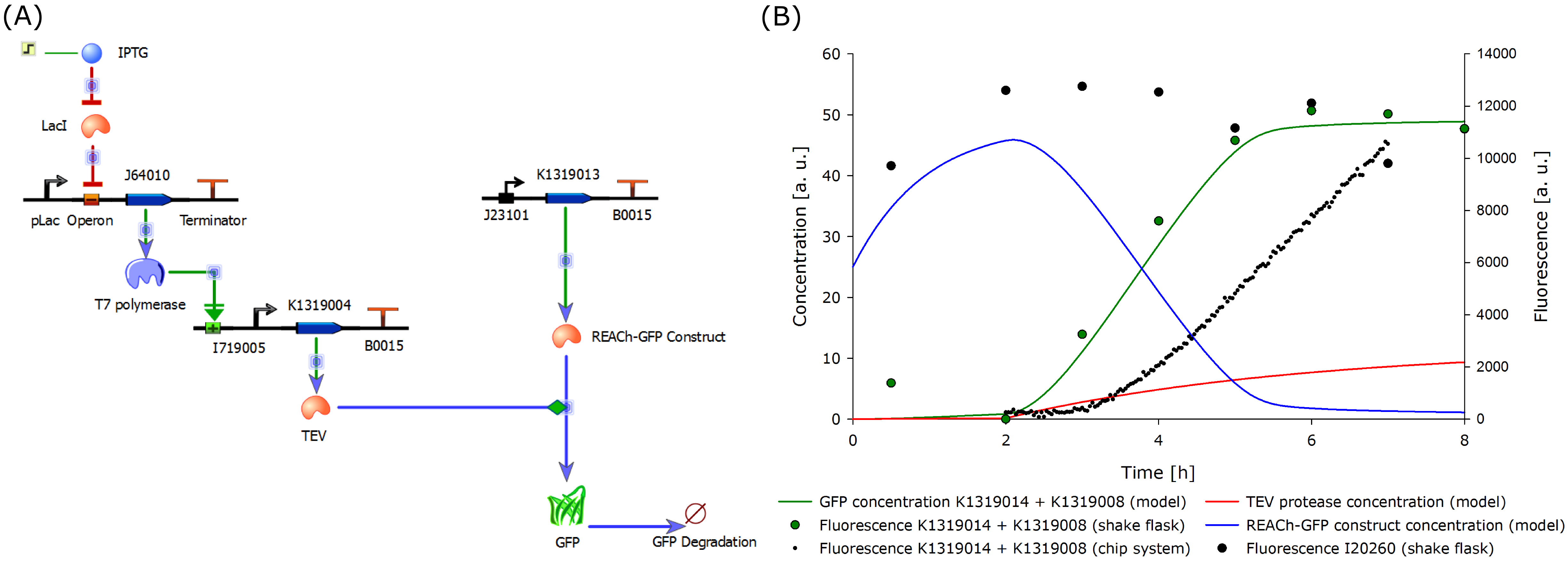
| - | The model was optimized to fit the data generated from the characterization experiment conducted in shake flasks. Additionally, the data from the characterization experiment of the double plasmid construct (K1319014 + K1319008) in the chip system was included in the plot above. The data was derived from the plate reader output of the four central spots of the chip. The background from the non-induced chip was substracted from the | + | The model was optimized to fit the data generated from the characterization experiment conducted in shake flasks. Additionally, the data from the characterization experiment of the double plasmid construct (K1319014 + K1319008) in the chip system was included in the plot above. The data was derived from the plate reader output of the four central spots of the chip. The background from the non-induced chip was substracted from the fluorescence response to correct the data and avoid effects from cell growth leading to wrong signal strengths. The development of the fluorescence is presented [https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#reachachievementschip here]. |
| - | It is shown that the | + | It is shown that the fluorescence response in chips develops later than in the characterization experiment in shake flasks. This is because the solid agar chip provides a higher diffusion barrier than liquid medium as used in the shake flasks. Further, a high oxygen transfer rate in shake flask cultivations enhances the rate of fluorescence development, as oxygen is needed for GFP production, in comparison to the cultivation in chips. |
| - | The model predictions correlate with the data generated from the characterization experiments, thus validating our molecular approach. With an iterative cycle of modeling, a faster and stronger | + | The model predictions correlate with the data generated from the characterization experiments, thus validating our molecular approach. With an iterative cycle of modeling, a faster and stronger fluorescence signal could be proven both theoretically and empirically. |
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
Revision as of 01:00, 18 October 2014
|
|
 "
"