Team:Oxford/biosensor characterisation
From 2014.igem.org
(Difference between revisions)
MatthewBooth (Talk | contribs) |
|||
| Line 101: | Line 101: | ||
<br><br> | <br><br> | ||
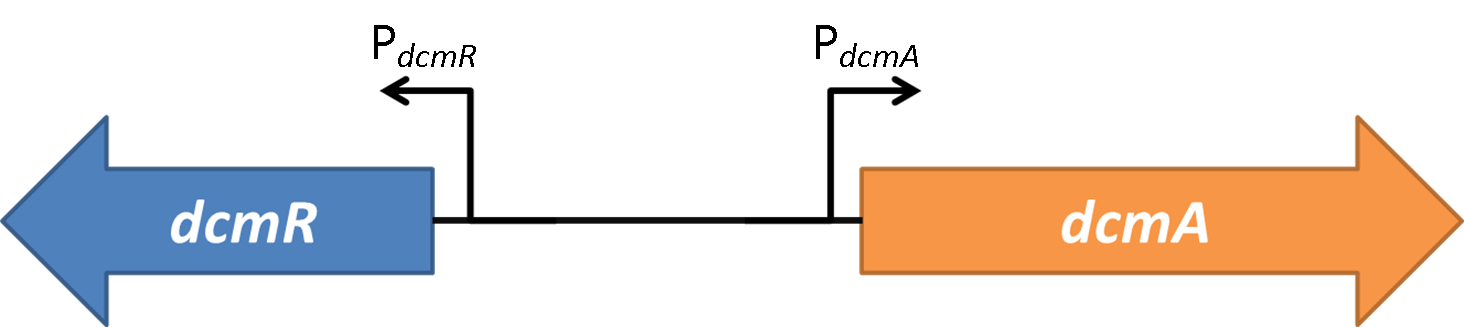
<h1>Characterising the DcmR - DCM - P_dcmA interaction</h1> | <h1>Characterising the DcmR - DCM - P_dcmA interaction</h1> | ||
| - | To find out whether the | + | To find out whether the DcmR acts as a repressor or an activator on the promoter of the <font style="font-style: italic;">dcmA</font> gene, we attempted to build the genetic circuit shown above on the right. Having <font style="font-style: italic;">dcmR</font> under inducible TetR expression should allow us to have very good control of the amount of DcmR present. Additionally a translational fusion with DcmR and a mCherry fluorescence tag will act as another confirmation to the amount of DcmR present. |
<br><br> | <br><br> | ||
We then extensively modelled the circuit to discover how the response of the system would differ if it was either of the two circuit systems. Click the modelling bubbles (pink) to find out exactly how we achieved this. | We then extensively modelled the circuit to discover how the response of the system would differ if it was either of the two circuit systems. Click the modelling bubbles (pink) to find out exactly how we achieved this. | ||
| Line 136: | Line 136: | ||
<h1>Predicting the mCherry fluorescence</h1> | <h1>Predicting the mCherry fluorescence</h1> | ||
| - | We simplified the first double repression by modelling it as an activation of <font style="font-style: italic;">dcmR</font> by ATC, albeit parameterised by different constants. This assumption is justified by the fact that we are able to precisely control the addition of ATC and measure the fluorescence of the mCherry. | + | We simplified the first double repression by modelling it as an activation of <font style="font-style: italic;">dcmR</font> by anhydrotetracycline (ATC), albeit parameterised by different constants. This assumption is justified by the fact that we are able to precisely control the addition of ATC and measure the fluorescence of the mCherry. |
<br> | <br> | ||
<br> | <br> | ||
| Line 195: | Line 195: | ||
As you can clearly see from the graph, the model predicts a large fluorescence increase as the input is added. This is the what we expect from the actual system and is the best approximation that is obtainable before we get experimental data. | As you can clearly see from the graph, the model predicts a large fluorescence increase as the input is added. This is the what we expect from the actual system and is the best approximation that is obtainable before we get experimental data. | ||
<br><br> | <br><br> | ||
| - | In the graph above, the model is set to have a basal transcription rate of zero. This is why there is a zero fluorescence response before the input has been added - this corresponds to the | + | In the graph above, the model is set to have a basal transcription rate of zero. This is why there is a zero fluorescence response before the input has been added - this corresponds to the Ptet promoter not being leaky. This basal rate will be calibrated alongside all of the other parameters in the model. |
</div> | </div> | ||
| Line 460: | Line 460: | ||
<h1>Wetlab data showing response in level of mCherry expressed with different concs of ATC</h1> | <h1>Wetlab data showing response in level of mCherry expressed with different concs of ATC</h1> | ||
| - | By making a translational fusion of mCherry at the C terminus of the dcmR gene under the tet promoter and tet operator system (see our <a href="https://2014.igem.org/Team:Oxford/biosensor_construction">Construction page</a> for details) we could measure mCherry fluorescence to gain information about dcmR induction by ATC. Expression was induced with various amounts of ATC and the following fluorescence data acquired. Exposure time was 0.2 seconds. As no calibration data was obtained using purified mCherry, the results have been left in fluorescence arbitrary units. Images were analysed using imageJ software.<br> | + | By making a translational fusion of mCherry at the C terminus of the dcmR gene under the tet promoter and tet operator system (see our <a href="https://2014.igem.org/Team:Oxford/biosensor_construction">Construction page</a> for details) we could measure mCherry fluorescence to gain information about <font style="font-style: italic;">dcmR</font> induction by ATC. Expression was induced with various amounts of ATC and the following fluorescence data acquired. Exposure time was 0.2 seconds. As no calibration data was obtained using purified mCherry, the results have been left in fluorescence arbitrary units. Images were analysed using imageJ software.<br> |
mCherry fluorescence increases with amount of ATC used confirming that the dcmR gene was expressed under the control of the tet promoter and operator system.<br><br> | mCherry fluorescence increases with amount of ATC used confirming that the dcmR gene was expressed under the control of the tet promoter and operator system.<br><br> | ||
<img src="https://static.igem.org/mediawiki/parts/5/51/Oxford_DcmR-mCherry_expression_induced_by_0ng_ATC.png" | <img src="https://static.igem.org/mediawiki/parts/5/51/Oxford_DcmR-mCherry_expression_induced_by_0ng_ATC.png" | ||
Revision as of 00:06, 18 October 2014



























 "
"








 Deterministic models are very powerful tools for synthetic biology. They describe the behaviour of the bacteria at the population level and use Ordinary Differential Equations (ODEs) to relate each activation and repression. By constructing a cascade of differential equations one can build a realistic model of the average behaviour of the system.
Deterministic models are very powerful tools for synthetic biology. They describe the behaviour of the bacteria at the population level and use Ordinary Differential Equations (ODEs) to relate each activation and repression. By constructing a cascade of differential equations one can build a realistic model of the average behaviour of the system.



 Oxford iGEM 2014
Oxford iGEM 2014














 This simplifies the equation to:
This simplifies the equation to:
 As we want our model to accurately predict the fluorescence, we will substitute the fluorescence value in place of the [DcmR] and rearrange:
As we want our model to accurately predict the fluorescence, we will substitute the fluorescence value in place of the [DcmR] and rearrange:
 Substituting in the value for δ1 that we found above and the basal steady state fluorescence level from the data (471 to 3 s.f.) gives the basal transcription rate as:
Substituting in the value for δ1 that we found above and the basal steady state fluorescence level from the data (471 to 3 s.f.) gives the basal transcription rate as:

 alongside the correct inputs:
alongside the correct inputs:
 The graph below shows the model's predictions plotted in the same figure as the data points that the wet-lab team obtained for the system:
The graph below shows the model's predictions plotted in the same figure as the data points that the wet-lab team obtained for the system:
 Plotting the model's output as a by interpolating between the calculated values makes the graph clearer:
Plotting the model's output as a by interpolating between the calculated values makes the graph clearer:


 The first system that we tested is shown here. This is the bottom level of our synthetic circuit. Testing just this plasmid allowed us to obtain some important information to allow us to characterise the genetic system.
The first system that we tested is shown here. This is the bottom level of our synthetic circuit. Testing just this plasmid allowed us to obtain some important information to allow us to characterise the genetic system.
 The second system that we tested was the whole synthetic system without any DCM added. This allowed us to analyse just the effect of DcmR on the PdcmA promoter when it was compared to the result from system 1.
The second system that we tested was the whole synthetic system without any DCM added. This allowed us to analyse just the effect of DcmR on the PdcmA promoter when it was compared to the result from system 1.
 Therefore we know that DcmR represses PdcmA with DCM repressing this repression. This means that the system is a double repressor!
Therefore we know that DcmR represses PdcmA with DCM repressing this repression. This means that the system is a double repressor!



































