Team:SUSTC-Shenzhen/Parts
From 2014.igem.org
(→Our RFC) |
(→Our RFC) |
||
| Line 47: | Line 47: | ||
<center>'''Fig.2 PCR Product Sequence'''</center> | <center>'''Fig.2 PCR Product Sequence'''</center> | ||
| - | The purpose to make such design because we do not know how many or what kind of bases interval between different promoters and polyAs will make the most transcription efficiency. Just like the data of [http://en.wikipedia.org/wiki/Kozak_consensus_sequence Kozak consensus sequence], we can measure it by changing the base numbers or kinds and finally find it. | + | The purpose to make such design because we do not know how many or what kind of bases interval between different promoters and polyAs will make the most transcription efficiency. Just like the data of ''[http://en.wikipedia.org/wiki/Kozak_consensus_sequence Kozak consensus sequence]'', we can measure it by changing the base numbers or kinds and finally find it. |
=='''Favorite Parts'''== | =='''Favorite Parts'''== | ||
Revision as of 23:10, 17 October 2014
Parts
Our devotion to the community, in reality.
Contents |
Our RFC
We made a new standard and we built three parts based on that this year. ([http://parts.igem.org/Part:BBa_K1431201 BBa_K1431201],[http://parts.igem.org/Part:BBa_K1431301 BBa_K1431301],[http://parts.igem.org/Part:BBa_K1431302 BBa_K1431302])
The BbsI restriction enzyme recognize the sequence GAAGAC and cut downstream two bases to six bases. If we make two reverse directions based on BbsI site, we can cut a part and insert another one with seamless. (Mechanism shown below)
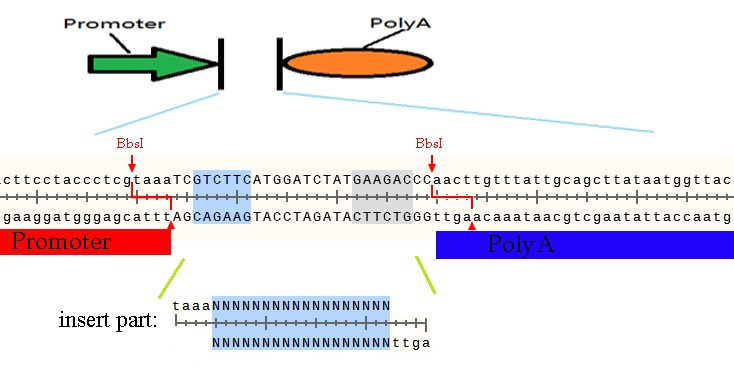
Let us make the above sequence as an example. If you want to insert a coding sequence between promoter and polyA, the prefix of forward primer you design must contain GAAGACNNtaaaNNNN and reverse must contain GAAGACNNagttNNNN. The middle two Ns would be any bases for them will be cut off and the four Ns should be begins or ends of the coding sequence. You can get parts by above primers and digest by BbsI restriction enzyme. Then you can insert the parts into promoter and polyA seamless. The PCR product must be such sequence, including the protect bases, shows below.
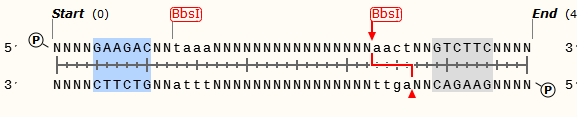
The purpose to make such design because we do not know how many or what kind of bases interval between different promoters and polyAs will make the most transcription efficiency. Just like the data of [http://en.wikipedia.org/wiki/Kozak_consensus_sequence Kozak consensus sequence], we can measure it by changing the base numbers or kinds and finally find it.
Favorite Parts

TRE 3G promoter + SV40 PolyA([http://parts.igem.org/Part:BBa_K1431301 BBa_K1431301])would be the best parts what we designed this year and also be our favorite parts.
Parts Table
 "
"