Team:UCLA/Project/Customizing Silk
From 2014.igem.org
Anuvedverma (Talk | contribs) |
Anuvedverma (Talk | contribs) |
||
| Line 110: | Line 110: | ||
<p dir="ltr"><sup><span>[2] </span></sup><span>Gatesy, John, et al. "Extreme diversity, conservation, and convergence of spider silk fibroin sequences." </span><em>Science</em><span> 291.5513 (2001): 2603-2605.</span></p> | <p dir="ltr"><sup><span>[2] </span></sup><span>Gatesy, John, et al. "Extreme diversity, conservation, and convergence of spider silk fibroin sequences." </span><em>Science</em><span> 291.5513 (2001): 2603-2605.</span></p> | ||
<p dir="ltr"><sup><span>[3] </span></sup><span>Xia, Xiao-Xia, et al. "Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber." </span><em>Proceedings of the National Academy of Sciences</em><span> 107.32 (2010): 14059-14063.</span></p> | <p dir="ltr"><sup><span>[3] </span></sup><span>Xia, Xiao-Xia, et al. "Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber." </span><em>Proceedings of the National Academy of Sciences</em><span> 107.32 (2010): 14059-14063.</span></p> | ||
| - | <p dir="ltr"><sup><span>[4] </span></sup><span>Huemmerich, Daniel, et al. "Novel assembly properties of recombinant spider dragline silk proteins." </span><em>Current Biology</em><span> | + | <p dir="ltr"><sup><span>[4] </span></sup><span>Huemmerich, Daniel, et al. "Novel assembly properties of recombinant spider dragline silk proteins." </span><em>Current Biology</em><span>;14.22 (2004): 2070-2074.</span></p> |
| - | <p dir="ltr"><sup><span>[5] </span></sup><span>Briggs, Adrian W., et al. "Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers." | + | <p dir="ltr"><sup><span>[5] </span></sup><span>Briggs, Adrian W., et al. "Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers."</span> <em>Nucleic acids research</em><span> (2012): gks624.</span></p> |
<p>[6] Teulé, Florence, et al. "A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning." <i>Nature protocols</i> 4.3 (2009): 341-355.</span></p> | <p>[6] Teulé, Florence, et al. "A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning." <i>Nature protocols</i> 4.3 (2009): 341-355.</span></p> | ||
<p><span> </span></p> | <p><span> </span></p> | ||
Revision as of 21:16, 17 October 2014

TEXT
TEXT
TEXT
TEXT
Customizing Silk
Background
Spider silk is an exceptional natural material. It exhibits remarkable strength and has the potential to be a very useful medium for a variety of applications. The properties of spider silk come primarily from the structure and amino acid sequence of the silk protein. One spider silk protein is comprised of repetitive units, referred to as monomers. Repeats of identical monomers make up one spider silk protein, and these proteins come together to form a silk fiber. The monomer is therefore the basic unit of the silk polypeptide, and changing the sequence alters the physical properties of the spider silk.[1]
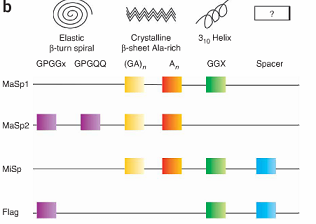
In fact, the sequence of the spider silk monomer differs slightly from species to species. Therefore, silk produced by different species have different physical properties. Some monomer sequences result in silks with greater strength, while other sequences result in silks with greater flexibility. Furthermore, individual spiders can produce multiple types of silk such as dragline and flagelliform silk, each with very different properties.[2]
Much research has been done on the production of spider silk from recombinant bacteria.[3] We want to build on this work and take it a step further. Using the wide range of spider silk monomers as our building blocks, we want to be able to create hybrid silk proteins with specified physical properties. By mixing and matching these different monomer sequences within one polypeptide, we hope to be able to fine tune the properties of our produced silk. If this system could be standardized, it could be exceedingly useful for quickly producing silk to fit different a variety of different needs and purposes.
For our project, we have chosen to start out with two different silk monomers, major ampullate spidroin 1 (MaSp1) and 2 (MaSp2). The MaSp1 monomer contributes strength to spider silk while the MaSp2 contributes more flexibility.[4] As a proof of principle we hope to show that by assembling these two repeats into one protein at different ratios, we can create silk with a spectrum of strength and extensibility.

Iterative Capped Assembly
Assembling spider silk monomers such as MaSp1 and MaSp2 together can prove to be rather difficult. Due to the repetitiveness of our silk monomers, common techniques such as Gibson assembly and direct synthesis of a full-length gene product could be ineffective and less efficient in building silk sequences. Therefore, a technique called iterative capped assembly (ICA), which allows rapid assembly of repeating monomers, would be a better option in working with our highly repetitive sequences. This technique has previously been applied in order to assemble other repetitive modules. For example, ICA has been demonstrated to efficiently assemble very repetitive transcription activator-like effector nucleases (TALENs) up to 21 monomers long[5].
ICA allows the assembly of silk monomers in different ratios and orders into a custom gene sequence of modifiable length. Gene monomers are assembled individually into a growing chain that is anchored to a solid foundation through streptavidin-biotin interaction.
Silk monomers for ICA were built using PCR to attach bsaI recognition site onto the MaSP sequences, as well as different end extensions in the form of 4bp overhang that is essential for ligation. BsaI is a type IIs endonuclease that cleaves outside the recognition site and therefore generates overhangs that are still part of the native silk sequence. This leaves our digested products, or building blocks, free of any remaining recognition site, which is usually formed with other types of restriction enzymes.

We have designed 3 types of overhangs that the Bsa1 enzyme can generate. These are the A overhang, (AGCA), the B overhang (TGCA) and the C overhang (TGCT).
The key idea here is that monomers with the same overhang are complementary and can be ligated together. Each silk monomer was modified via pcr to have one of these overhangs at the 5’ end of the sequence and another at the 3’ end of the sequence. One version of our silk monomer had an A overhang at the 5’ end and a B overhang at the 3’ end, which we called (5’)AB(3’). Another version was (5’)BC(3’), and the final version was (5’)CA(3’). We therefore end up with 3 versions of the same silk monomer. Making these modifications for all of our different types of silk monomers potentially gives us the ability to assemble a hybrid silk gene composed of different monomers in a matter of hours. Following restriction digestion of all our monomers with BsaI to create the sticky ends, each silk monomer would be able to ligate to the preceding piece due to the complementing 4bp on their ends. For example, if we wished to ligate a MaSp 2 monomer to a (5’)BC(3’)MaSp1 monomer, we would simply add a (5’)CA(3’) MaSp2. With the 3 subsets of MaSP1 and MaSP2, we could eventually program gene sequence of desired physical properties with various ratios and orders of each monomer type.
A key aspect of ICA is that the gene to be assembled is fixed to a solid support as it is being ligated together. Streptavidin beads act as the solid support in this case. Conjugating our gene to the beads therefore necessitates an “initiator oligo”, a biotynilated sequence that contains both the biobrick prefix as well as one of the A,B, or C overhangs at its 3’ end. An advantage of fixing our growing sequence to the beads is that it allows us to remove all traces of the previous ligation in a “wash” step before adding the next silk monomer to the growing gene.

When dealing with repetitive sequences, regular assembly techniques would give products of various lengths due to the uncontrolled ligation among pieces. Another powerful aspect of ICA is that it increases the frequency of producing a full-length sequence that we intend to build. This is achieved by adding capping oligos that ligate to a chain where a previous monomer piece fail to attach. Any incomplete or incorrect sequence due to unsuccessful ligations would be blocked from further extending, and thus monomers that are added later would ligate to the right sequences at a higher frequency.
Once the full gene sequence has been generated, the terminator oligo containing the biobrick suffix is added to complete the assembly. The gene can then be released from the streptavidin beads by heating and disrupting the Streptavidin biotin interaction[5].
ICA is a proficient technique to standardize assembly of any custom gene sequence. Not only does it provide flexibility in producing custom gene sequence, it is also compatible with the iGEM biobrick system. The initiator contains the prefix sequence of a biobrick and the terminator contains the suffix sequence. These contain primer sites allow PCR amplification of the full-length constructs after the release from the beads. In addition, restriction sites within the initiator and terminator enable the insertion of the complete, assembled product into a biobrick backbone easily using Golden-Gate cloning.
What We've Done
Over the course of the summer, we modified the nucleotide sequence of MaSp1 and MaSp2 so that it is both compatible with the iGEM standards as well as with our ICA assembly scheme. We generated these sequences by ordering single stranded oligos and doing multiple rounds of pcr to assemble the full construct. These sequences contain the biobrick prefix and suffix as well as the required Bsa1 restriction sites for the ICA/Golden Gate reaction. Furthermore, we designed and assembled the other nucleotide sequences necessary for ICA from primers that we ordered. These sequences include the biotinylated initiator oligo, the ICA terminator oligo, as well as the capping oligos (see above for description of each).
We ran some trial runs of ICA by trying to assemble only a few monomers together. We only were able to run a few trials, and unfortunately, were not able to create the full silk construct. We were able to ligate the initiator oligo to the terminator oligo, showing that the initiator oligo was successfully conjugating to the streptavidin beads and that the ligation reaction reaction worked. Hopefully with some optimizations and troubleshooting, we can use this protocol to assemble a full length silk construct in the near future.
References
[1] Lewis, Randolph V. "Spider silk: ancient ideas for new biomaterials." Chemical reviews 106.9 (2006): 3762-3774.
[2] Gatesy, John, et al. "Extreme diversity, conservation, and convergence of spider silk fibroin sequences." Science 291.5513 (2001): 2603-2605.
[3] Xia, Xiao-Xia, et al. "Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber." Proceedings of the National Academy of Sciences 107.32 (2010): 14059-14063.
[4] Huemmerich, Daniel, et al. "Novel assembly properties of recombinant spider dragline silk proteins." Current Biology;14.22 (2004): 2070-2074.
[5] Briggs, Adrian W., et al. "Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers." Nucleic acids research (2012): gks624.
[6] Teulé, Florence, et al. "A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning." Nature protocols 4.3 (2009): 341-355.
 "
"
