Team:Austin Texas/kit
From 2014.igem.org
| Line 244: | Line 244: | ||
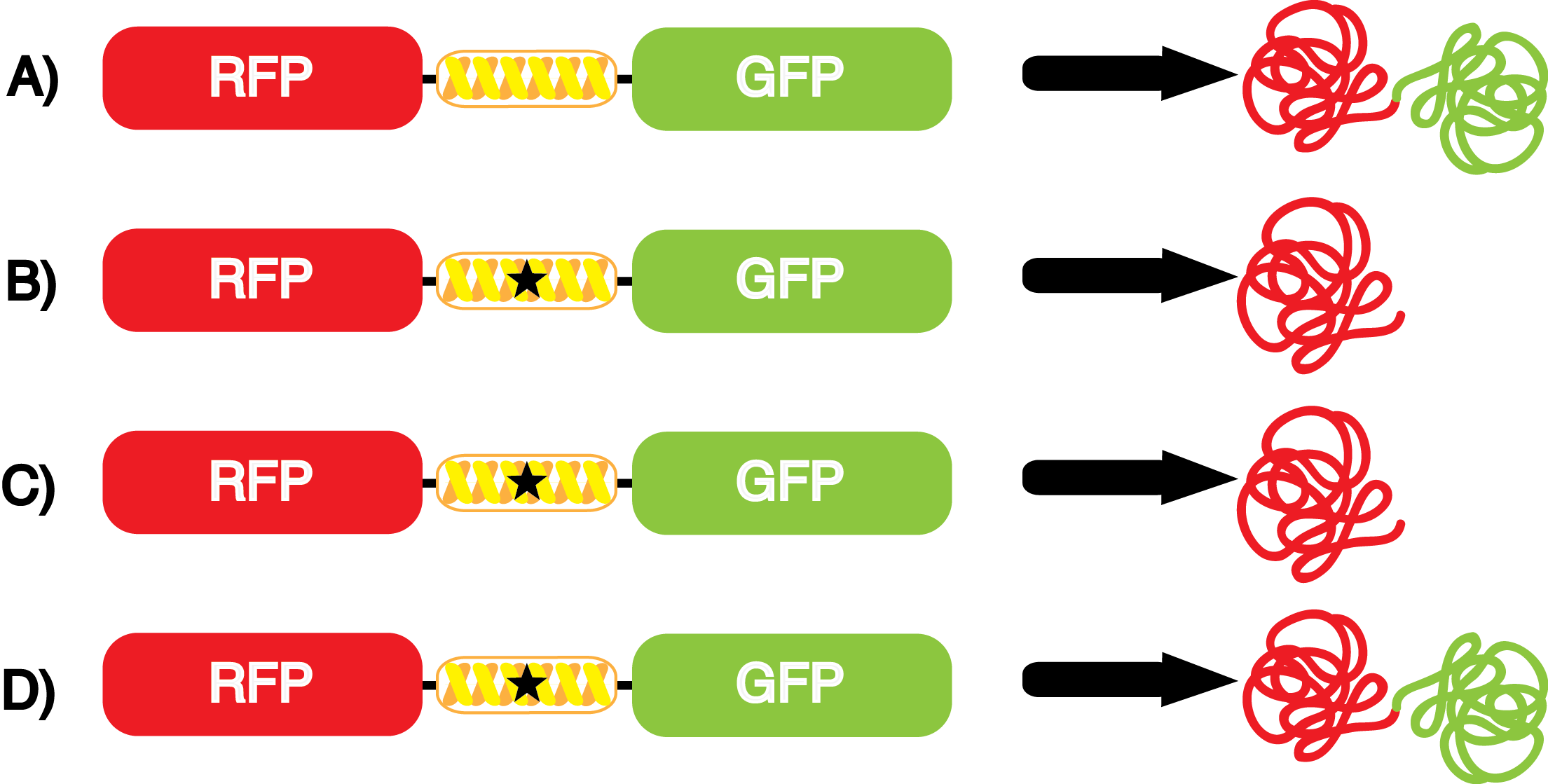
This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition by contributing to ncAA methods of analysis and fulfilling [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Our team has successfully designed a unique testing kit for ncAA incorporation <i>in vivo</i> using common reporter parts to characterize very uncommon synthetase/tRNA machinery. In addition, we have used our kit to analyze and characterize seven different ncAA synthetase/tRNA pairs as well as tyrosine RS. This method of testing was designed to be undergraduate administrable, transportable, and more affordable than other methods of characterization and analysis. | This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition by contributing to ncAA methods of analysis and fulfilling [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Our team has successfully designed a unique testing kit for ncAA incorporation <i>in vivo</i> using common reporter parts to characterize very uncommon synthetase/tRNA machinery. In addition, we have used our kit to analyze and characterize seven different ncAA synthetase/tRNA pairs as well as tyrosine RS. This method of testing was designed to be undergraduate administrable, transportable, and more affordable than other methods of characterization and analysis. | ||
| + | |||
| + | Documentation, including sequence information, of the tRNA synthetase/tRNA pairs used can be found [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Synthetase_Table below]. | ||
| + | |||
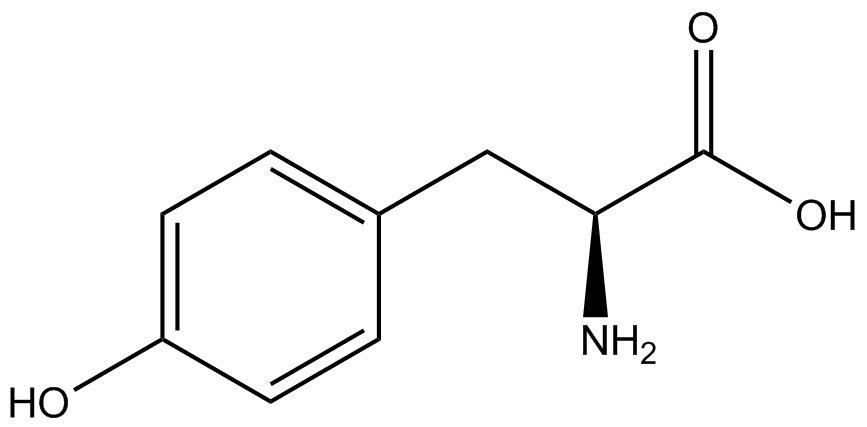
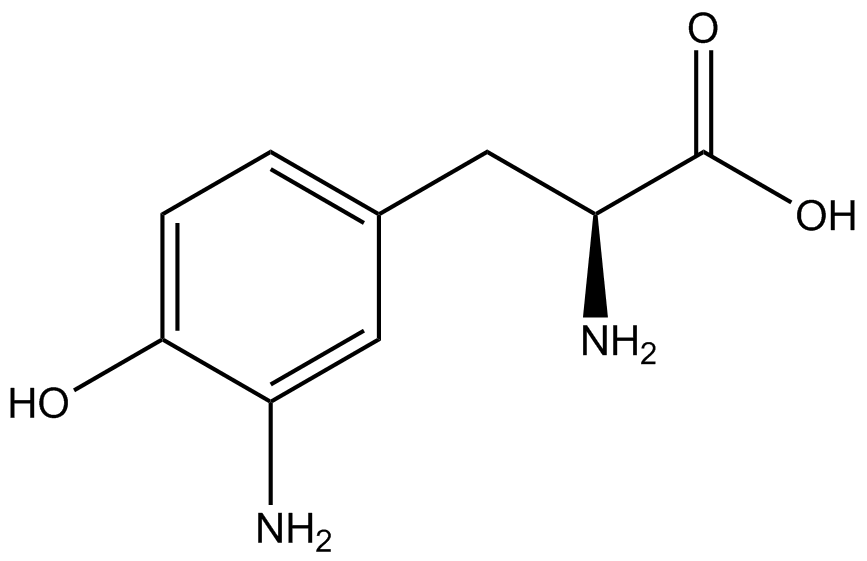
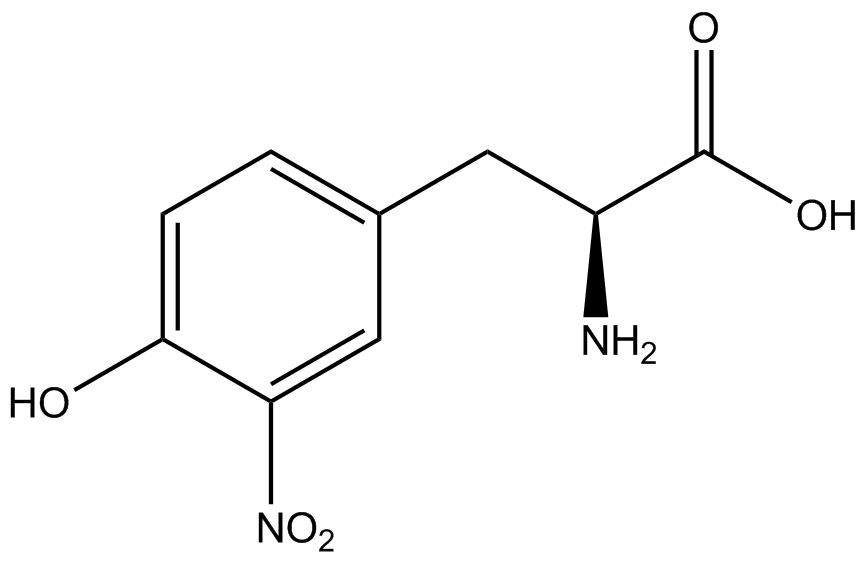
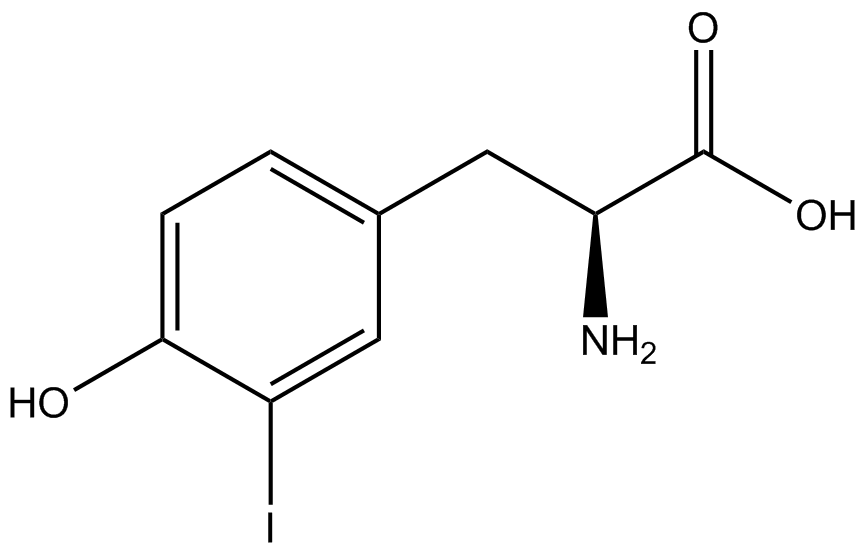
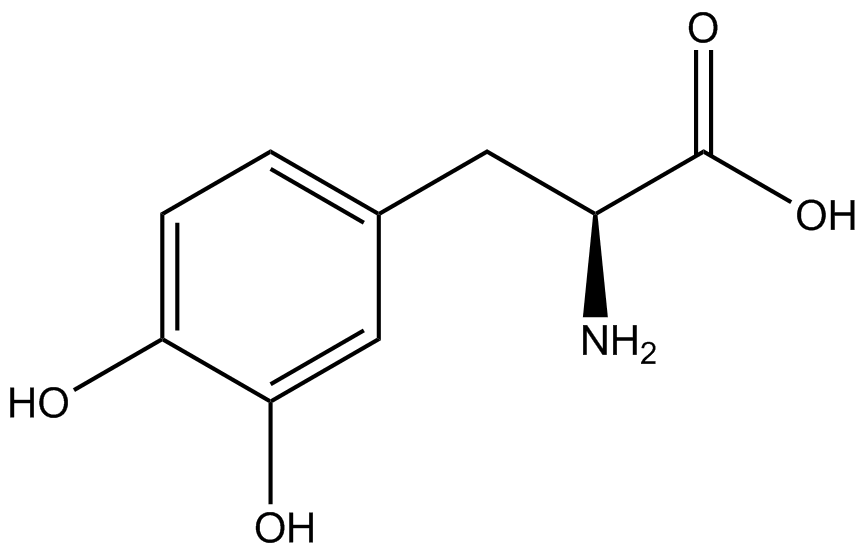
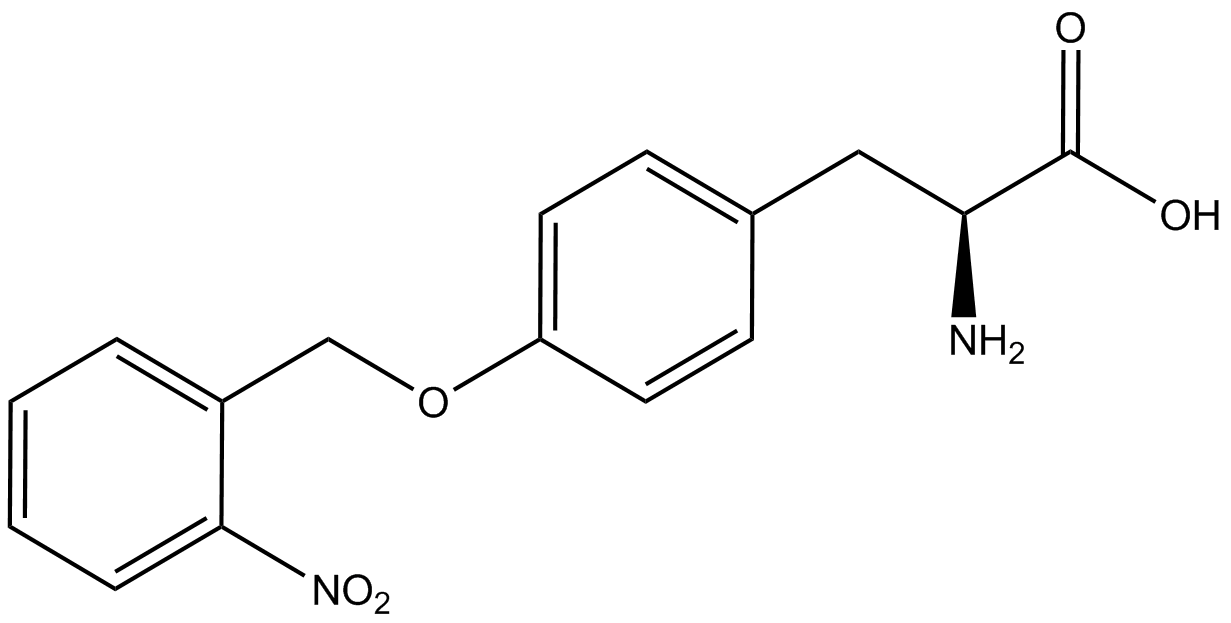
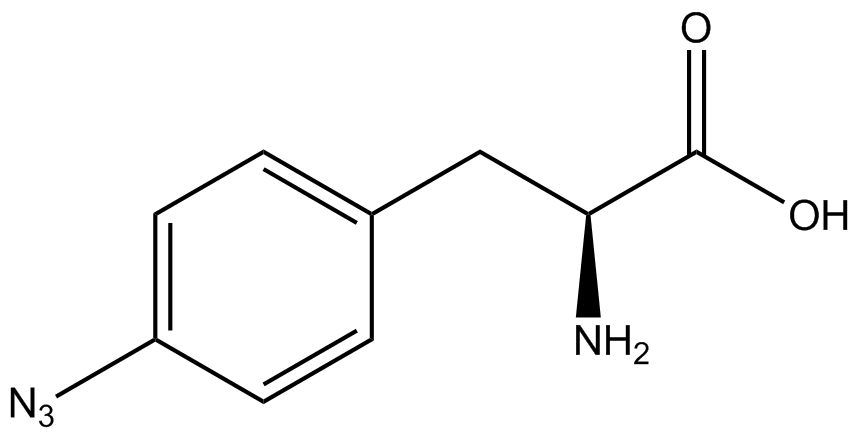
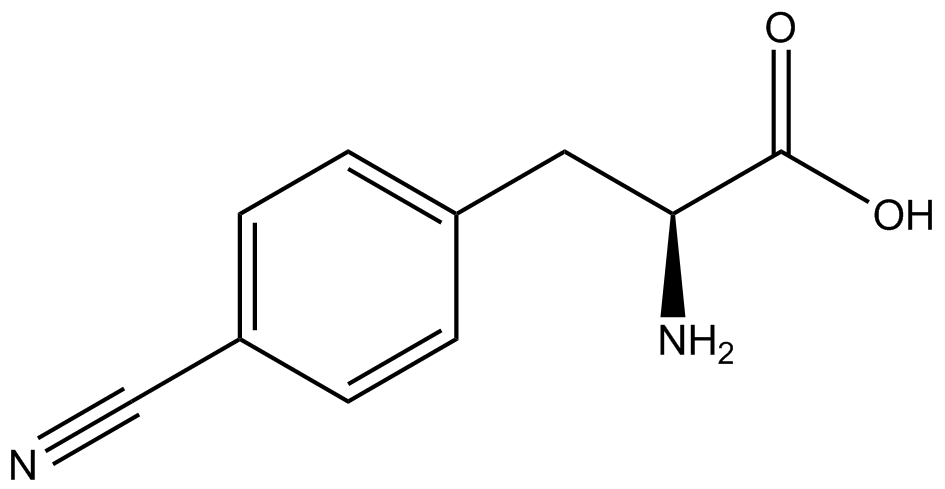
| + | Information regarding the ncAA used can also be found [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table below]. | ||
<h1>ncAA Table</h1> | <h1>ncAA Table</h1> | ||
Revision as of 18:01, 17 October 2014
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"