Team:Lethbridge/results
From 2014.igem.org
| Line 86: | Line 86: | ||
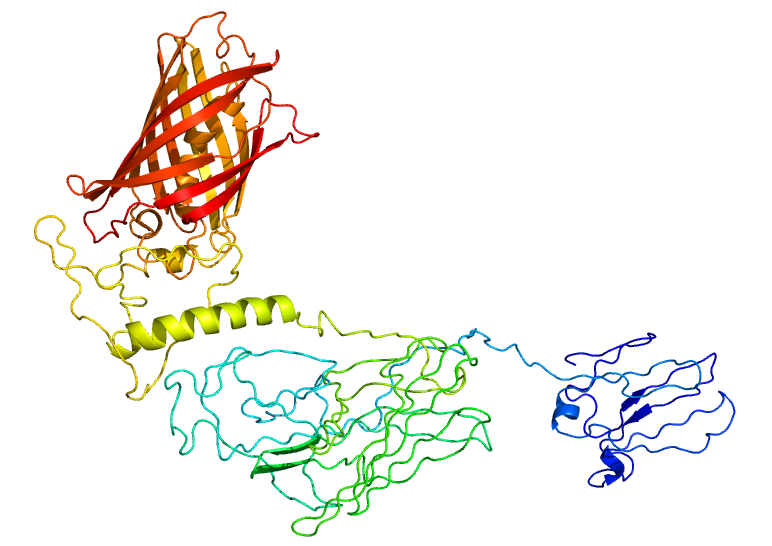
<p>To test if our RVG-Lamp2B construct was being appropriately expressed and folded, we fused Clover (a green fluorescent protein) to the C-terminus of Lamp2B. Using the Phyre2 server, we were able to generate a model of the predicted structure of our RVG-Lamp2B-Clover fusion protein based on homology and multi-template <i>ab initio</i> modeling.</p> | <p>To test if our RVG-Lamp2B construct was being appropriately expressed and folded, we fused Clover (a green fluorescent protein) to the C-terminus of Lamp2B. Using the Phyre2 server, we were able to generate a model of the predicted structure of our RVG-Lamp2B-Clover fusion protein based on homology and multi-template <i>ab initio</i> modeling.</p> | ||
| - | <center>[[Image:Lamp2B_Clover_Structure.jpg| | + | |
| + | <center>[[Image:Lamp2B_Clover_Structure.jpg|500px]]</center> | ||
<p><b>Figure 1: The predicted structure of the RVG-Lamp2B-Clover fusion protein.</b> The Clover domain is obvious in red with Lamp2B below in green and the RVG domain evident at the bottom in blue.</p><br> | <p><b>Figure 1: The predicted structure of the RVG-Lamp2B-Clover fusion protein.</b> The Clover domain is obvious in red with Lamp2B below in green and the RVG domain evident at the bottom in blue.</p><br> | ||
| Line 100: | Line 101: | ||
| - | <center>[[Image:Fluorimeter_data.jpg| | + | <center>[[Image:Fluorimeter_data.jpg|700px]]</center> |
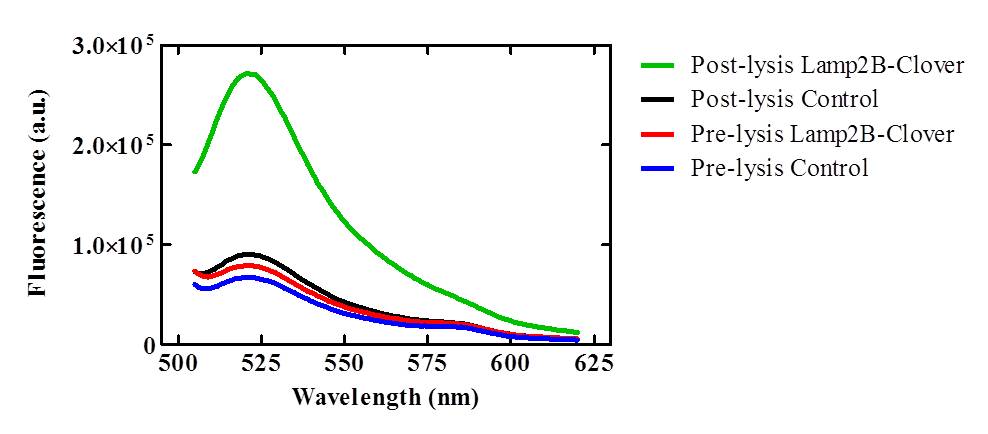
<p><b>Figure 3: Lysed exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected HEK-293 cells demonstrate elevated fluorescence in the 505-620nm range relative to control and unlysed samples.</b> Description goes here.</p><br> | <p><b>Figure 3: Lysed exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected HEK-293 cells demonstrate elevated fluorescence in the 505-620nm range relative to control and unlysed samples.</b> Description goes here.</p><br> | ||
| Line 111: | Line 112: | ||
<p>To test if expression of NeuroD1 has a phenotypic effect on astrocytes, pcDNA3.0-NeuroD1-Clover was lipofected into (or just added to the media of) C8-D30 cultures. After one week, all astrocyte cells demonstrated cytosolic Clover expression and very weak nuclear expression of NeuN, an indicator of mature neurons. However, the cell bodies and processes were still astrocytic in appearance indicating either an insufficient level of NeuroD1 expression to induce changes to a neural phenotype OR a problem with the design/construction of the plasmid. We are currently addressing both of these possibilities.</p> | <p>To test if expression of NeuroD1 has a phenotypic effect on astrocytes, pcDNA3.0-NeuroD1-Clover was lipofected into (or just added to the media of) C8-D30 cultures. After one week, all astrocyte cells demonstrated cytosolic Clover expression and very weak nuclear expression of NeuN, an indicator of mature neurons. However, the cell bodies and processes were still astrocytic in appearance indicating either an insufficient level of NeuroD1 expression to induce changes to a neural phenotype OR a problem with the design/construction of the plasmid. We are currently addressing both of these possibilities.</p> | ||
| + | |||
<center>[[Image:Astrocyte_Clover_NeuN.JPG|700px]]</center> | <center>[[Image:Astrocyte_Clover_NeuN.JPG|700px]]</center> | ||
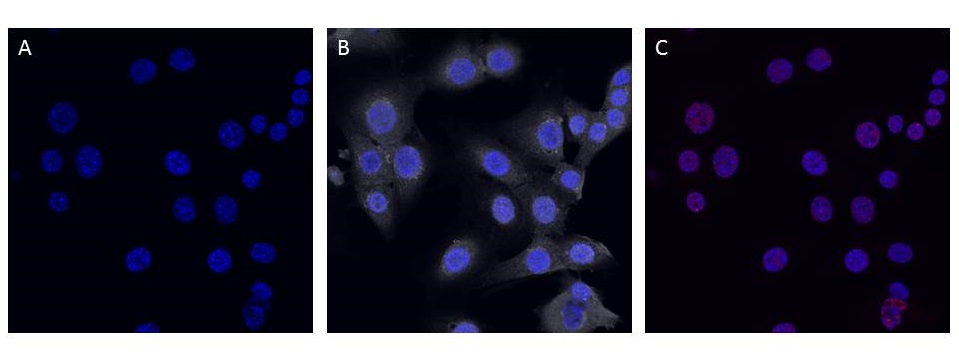
<p><b>Figure 4: C8-D30 (astrocyte) cell cultures transfected with pcDNA3.0-NeuroD1-Clover demonstrate elevated cytosolic Clover and weak nuclear NeuN expression.</b> Description goes here.</p><br> | <p><b>Figure 4: C8-D30 (astrocyte) cell cultures transfected with pcDNA3.0-NeuroD1-Clover demonstrate elevated cytosolic Clover and weak nuclear NeuN expression.</b> Description goes here.</p><br> | ||
Revision as of 01:15, 17 October 2014

Contents
Results
Team Parts Sandbox
Name Type Description Designer Length Favorite Part [http://parts.igem.org/Part:BBa_K1419000 BBa_K1419000] Device Arabinose inducible lysis casette Zak Stinson 3002 [http://parts.igem.org/Part:BBa_K1419001 BBa_K1419001] RNA Part Arabinose inducible lysis casette with RNA-IN Zak Stinson 32 [http://parts.igem.org/Part:BBa_K1419002 BBa_K1419002] Device RNA-OUT component for an Arabinose inducible lysis casette Harland Brandon 117****#$*#$ [http://parts.igem.org/Part:BBa_K1419003 BBa_K1419003] Device RNA-OUT component for a Universal Regulation System Harland Brandon 117****#$*#$ [http://parts.igem.org/Part:BBa_K1419004 BBa_K1419004] Device TEV Protease Harland Brandon 729DFDGFHFDHF [http://parts.igem.org/Part:BBa_K1419005 BBa_K1419005] Device Lamp2B-Clover Harland Brandon 2241 Yes Expression and localization of RVG-Lamp2B protein construct
To test if our RVG-Lamp2B construct was being appropriately expressed and folded, we fused Clover (a green fluorescent protein) to the C-terminus of Lamp2B. Using the Phyre2 server, we were able to generate a model of the predicted structure of our RVG-Lamp2B-Clover fusion protein based on homology and multi-template ab initio modeling.
Figure 1: The predicted structure of the RVG-Lamp2B-Clover fusion protein. The Clover domain is obvious in red with Lamp2B below in green and the RVG domain evident at the bottom in blue.
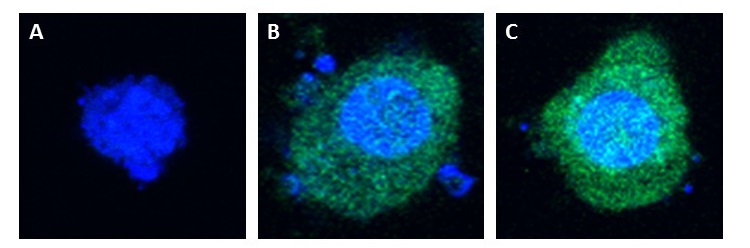
The pcDNA3.0-RVG-Lamp2B-Clover plasmid and a pcDNA3.0-Clover control were lipofected into separate Human Embryonic Kidney (HEK-293) cell cultures (HEK-293 cells are a resilient and rapidly dividing cell line also known to produce exosomes). In both lipofected cultures, there was elevated cytosolic green fluorescence relative to non-lipofected controls, which confirms expression and proper folding of the protein construct.
Figure 2: Confocal (60x) images of non-lipofected control HEK-293 cells versus pcDNA3.0-RVG-Lamp2B-Clover and pcDNA3.0-Clover lipofected HEK-293 cells. Nuclei were counterstained with DAPI.
To confirm that the RVG-Lamp2B-Clover protein construct was being targeted to exosomal membranes (and were not simply cytosolic), culture media was collected from the lipofected and control HEK-293 cell cultures and exosomes were purified via ultracentrifugation. After lysis, the exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected cultures demonstrated an elevated level of fluorescence (505-620nm) that corresponds to the emission wavelength of Clover (515nm) indicating appropriate targeting of the engineered protein to exosomal membranes.
Figure 3: Lysed exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected HEK-293 cells demonstrate elevated fluorescence in the 505-620nm range relative to control and unlysed samples. Description goes here.
We are now in the process of replacing Clover with a Transcription Activator-Like Effector (TALE) DNA-binding protein to the C-terminus of Lamp2B to anchor the therapeutic NeuroD1 plasmid within newly formed exosomes.
Astrocytic expression of NeuroD1 and resulting phenotype
To test if expression of NeuroD1 has a phenotypic effect on astrocytes, pcDNA3.0-NeuroD1-Clover was lipofected into (or just added to the media of) C8-D30 cultures. After one week, all astrocyte cells demonstrated cytosolic Clover expression and very weak nuclear expression of NeuN, an indicator of mature neurons. However, the cell bodies and processes were still astrocytic in appearance indicating either an insufficient level of NeuroD1 expression to induce changes to a neural phenotype OR a problem with the design/construction of the plasmid. We are currently addressing both of these possibilities.
Figure 4: C8-D30 (astrocyte) cell cultures transfected with pcDNA3.0-NeuroD1-Clover demonstrate elevated cytosolic Clover and weak nuclear NeuN expression. Description goes here.
 "
"