Team:HNU China/Project
From 2014.igem.org
(Difference between revisions)
Yannickchou (Talk | contribs) |
|||
| Line 461: | Line 461: | ||
</div> | </div> | ||
<div id="cont_5"> | <div id="cont_5"> | ||
| - | <div id="chead"></div> | + | <div id="chead"> |
| - | <div id="cmiddle"> | + | <div align="left"class="STYLE3" style="margin-left:50px;"><br/><br/><br/>Future work</div> |
| + | </div> | ||
| + | <div id="cmiddle"> | ||
| + | <div align="left" class="STYLE2" style="margin-left:50px;margin-right:50px;"> | ||
| + | <b>Step I. </b><br><br> | ||
| + | |||
| + | When all of the schematic work finished, we are firstly going to verify the hypothesis of the apoptosis effect when caspase-3 expressed in yeast.<br><br> | ||
| + | |||
| + | <b>Step II. <b><br><br> | ||
| + | |||
| + | The kinetic parameter of the light mediation, iron sensing and iron absorption are with worth to detect.<br><br> | ||
| + | |||
| + | <b>Step III.<b><br><br> | ||
| + | |||
| + | Though we would like to see them in only one microorganism clone, according to the original design, iron sensitive absorbing system and optogenetic apoptosis system consists of two engineered plasmids repectively. Four plasmids in total makes it impossible for integration. Further genetically engineering design and operation are necessary, mainly in system simplification. | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
<div id="cfoot"></div> | <div id="cfoot"></div> | ||
</div> | </div> | ||
Revision as of 23:43, 17 October 2014
Background
As far as the bronze age, our ancestors began to extract valuable metal from the mineral, with the combined process of chemical reaction and high temperature treatment, using quite a lot energy. When this ancient industry comes to the modern time, piles of problems emerge, then hinder the road to a more industrialized life, both more comfortable and material-consuming, some of which show as below:
- Due to the long history of mineral exploitation, people look for high purity reserves deep underneath the surface, DANGEROUS, making it to the miners.
- HIGH UTILIZATION RATIO is what we are appealing to the mineral waste slag as well the abandoned mines, which abound with reserves, however is not cost-effective under the treatment of traditional way.
- Besides of the slag, metal reserves shall also exist in the CIVIL WASTE. since electronic device used and threw in our daily life. Trash leachate is another object we resort to collect the valuable metal.
Description
Our microbial miner briefly consists of two systems, iron sensitive absorbing system and optogenetic apoptosis system, the last of which is designed for the biosafety reason.
- Iron sensitive absorbing system:
- IRE: part of the upstream none-coding sequence of human ADH1 RNA, which will form a hairpin structure with human protein IREBP, inhibiting the process of translation.
- IREBP: human protein.
- FET3: baker’s yeast protein, plays a substaintial role in the iron uptake.
- optogenetic apoptosis system
- Cry2-CIB1: both of these transcription factors are cloned from the genome of Arabidopsis, which will react under the blue light, then initiate the expression of the gene downstream.
- Casp3: human protein, plays a core role in the cellular apoptosis.
Experimentation
Preliminary construction of the Cry2-CIB1 system
I.Background
Photoactivation mechanism of Arabidopsis cryptochrome 2(CRY2), in mediating light regulation of cell elongation and photoperiodic flowering, has been revealed as a blue light-specific manner cooperating with cryptochrome-interacting basic helix-loop-helix protein(CIB1), which is screened by yeast two-hybrid assay.[1] This system is recently engineered in a light-inducible transcriptional effectors (LITEs), applying in mice neuron or the brain in vivo to mediate reversible gene expression.[2] Reasonably, we would like to utilize them to serve for our project, which in detail is an optogenetic apoptosis system for the biosafety reason.
II.Cloning of the Cry2 & CIB1 gene
According to the sequences from NCBI, we designed two pairs of primers respectively, and cloned them based on the cDNA library of Arabidopsis.[fig.1]
Primer for Cry2:
Fwd 5’→3’ ATGAATGGAGCTATAGGA
Rev 5’→3’ TCAAACTCCTAAATTGCC
Primer for CIB1:
Fwd 5’→3’ ATGAAGATGGACAAAAAGA
Rev 5’→3’ TCATTTGCAACCATTTTT
III.Optogenetic yeast-two-hybrid
Two-hybrid or interaction trap systems exploit the fact that transcription factors are comprised of two domains, a DNA binding domain (DBD) and an activation domain (AD). Two separate hybrid proteins are constructed in two-hybrid screens. The first hybrid protein is the DBD/protein X, while the second hybrid protein is the AD/protein Y.These two hybrids are encoded on separate yeast expression plasmids, with independent selectable markers.[3] Based on the principle of yeast-two-hybrid, there are several pairs of plasmids designed for the screening of interacting protein library, commercially accessible.
What we chose is the pDEST32 and pDEST22[fig.2], where the DBD and AD is actually Gal4 BD and Gal4 AD respectively, and the Gal4 AD shall specifically bind to the upstream activating sequence(UAS), locating in the promoters of several reporter genes in the yeast strain genome like lacZ, HIS3 as well URA3.
Fig.2

Since we have obtained CRY2 and CIB1, what we do next is to induce both of them into their vectors with the technology named Gateway, constructing two new plasmids,
pDEST22-CIB1 and pDEST32-32 as below[fig.3]:
Fig.3

Then we transform the plasmids above into the baker’s yeast strain, AH109, screening by Leu and Trp auxotroph. To test the effect of light switchable gene expression system, blue-white selection is induced when the blue light in special incubator[fig.4] shall initiate the expression of LacZ, which combined with the IPTG and X-gal in solid medium, makes the colony into blue, contrarily, colony in dark remains white.[fig.5]
Fig.4

Reference
- Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis, Hongtao Liu, Xuhong Yu, Kunwu Li, John Klejnot, Hongyun Yang, Dominique Lisiero, Chentao Lin*, SCIENCE VOL 322 5 DECEMBER 2008
- Optical control of mammalian endogenous transcription and epigenetic states, Silvana Konermann1,2*, Mark D. Brigham1,2,3*, Alexandro Trevino1,2, Patrick D. Hsu1,2,4, Matthias Heidenreich1,2, Le Cong1,2,5, Randall J. Platt1,2, David A. Scott1,2, George M. Church1,6 & Feng Zhang1,2, doi:10.1038/nature12466
- ProQuest Two-Hybrid System user manual.
Cloning of the genes we need and Construction of T-vector
I.IRE-IRP
The regulation of the synthesis of ferritin in mammalian cells is mediated by the interaction of the Iron regulatory protein (IRP) with a specific recognition site, the iron responsive element (IRE), In the 5' untransiated regions (UTRs) of the respective mRNAs. [1]
1.IRP
We decide to clone the IRP from human cDNA library.
Primers for IRP[fig.1]:
Fwd 5’→3’ GCGAAGCTTTCAGTAATCATGAGCAAC
HindIII
Rev 5’→3’ GCGGAGCTCTTGAGCAGAGCGTAAGA
SacI
Fig.1

2.IRE
Since IRE is a quite short sequence, counting 40 bp at all. A pair of single-stranded DNA are synthesized as the primers, and annealing shall create double-stranded DNA.
Pair of single-stranded DNAs
Fwd 5’→3’ GCGAAGCTTGTTCTTGCTTCAACAGTGTTTGGACGGAACACTAGTGCG
HindIII SpeI
Rev 5’→3’ CGCACTAGTGTTCCGTCCAAACACTGTTGAAGCAAGAACAAGCTTCGC
SpeI HindIII
II.FET3
S. cerevisiae accumulate iron by a process requiring a ferrireductase and a ferrous transporter. [2] FET3 is an oxidase which depends on the Cu2+ in yeast membrance, and forms the complex for iron absorption with FTR1, which is the main part of the high absorption system[3]. Here we hope FET3 could active the function of high absorption system to make the system more efficient in absorption of iron.
1.Prepare for the genome of baker’s yeast
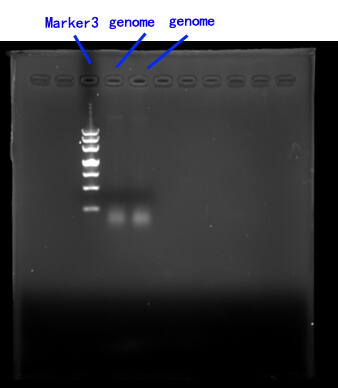
Due to the lack of existing yeast genome as the PCR template, we order a yeast genome extraction kit to prepare the source.[fig.2]
Fig.2
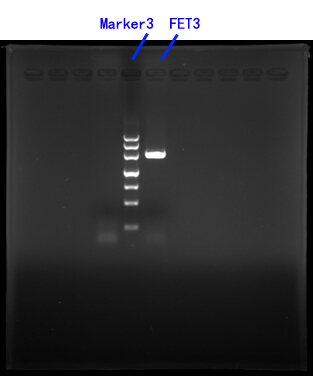
2.Cloning of FET3
Primers for FET3[fig.3]
Fwd 5’→3’ GCGAAGCTTATGACTAACGCTTTGCTCTCTATAG
HindIII
Rev 5’→3’ GCGGAGCTCTGGAACCCTTGACCGA
SacI
Fig.3
III.CASP3
Caspase-3 normally exists in the cytosolic fraction of cells as an inactive precursor that is activated proteolytically when cells are signaled to undergo apoptosis.[4] So in the project, we want to make the modified yeast commit suicide through overexpress caspase-3 protein to protect natural environment.
The template for PCR is also the human cDNA library.
Primers for CASP3[fig.4]
Fwd 5’→3’ GCGACTAGTGCTCTGGTTTTCGGTGGG
Spe1
Rev 5’→3’ GCGGAGCTCTGGAACCCTTGACCGA
SacI
Fig.4

IV.Construction of the T-vector
To make sure the PCR-cloned sequences are expected, we ligate them into the pMD18-T vector, then sequence all together. On the other hand, objective sequence in circular plasmids makes it convenient and stable for storage for quite long time.
Reference
- Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae, Carla C.Oliveira, Britta Goossen1, Nilson l.T.Zanchin, John E.G.McCarthy*, Matthias W.Hentze1 and Renata Stripecke1, Nucleic Acids Research, 1993, Vol. 21, No. 23
- The FET3 gene of S. cerevisiae encodes a multicopperoxidase required for ferrous iron uptake, Askwith, C., Eide, D., Van Ho, A., Bernard, P.S., Li, L., Davis-Kaplan, S., Sipe, D.M., Kaplan, J. Cell(1994)
- Stearman R, Yuan D S, Yamaguchi-Jwai Y et al. Science, 1996, 271(15): 1552~1557
- Activation of cyclin A-dependent protein kinases during apoptosis. Proc. Natl. Acad. Sci. USA, 91 (1994), pp. 3754–3758
4
Future work
Step I.
When all of the schematic work finished, we are firstly going to verify the hypothesis of the apoptosis effect when caspase-3 expressed in yeast.
Step II.
The kinetic parameter of the light mediation, iron sensing and iron absorption are with worth to detect.
Step III.
Though we would like to see them in only one microorganism clone, according to the original design, iron sensitive absorbing system and optogenetic apoptosis system consists of two engineered plasmids repectively. Four plasmids in total makes it impossible for integration. Further genetically engineering design and operation are necessary, mainly in system simplification.
When all of the schematic work finished, we are firstly going to verify the hypothesis of the apoptosis effect when caspase-3 expressed in yeast.
Step II.
The kinetic parameter of the light mediation, iron sensing and iron absorption are with worth to detect.
Step III.
Though we would like to see them in only one microorganism clone, according to the original design, iron sensitive absorbing system and optogenetic apoptosis system consists of two engineered plasmids repectively. Four plasmids in total makes it impossible for integration. Further genetically engineering design and operation are necessary, mainly in system simplification.
 "
"







