Team:SUSTC-Shenzhen/Notebook/Parts building
From 2014.igem.org
| Line 110: | Line 110: | ||
<span style="color: red">''<u>PCR TetOn, PB5, PB3, SV40/CBh/pTRE-3G promoter, PolyA1, 2*UAS</u>'' | <span style="color: red">''<u>PCR TetOn, PB5, PB3, SV40/CBh/pTRE-3G promoter, PolyA1, 2*UAS</u>'' | ||
| - | For TetOn and CBh were more than 700bp, we decided to use Q5® High-Fidelity DNA Polymerases, but others by OneTaq® 2X Master Mix. 2*UAS without template because the forward primer and reverse primer coμld combine each other and extend. | + | For TetOn and CBh were more than 700bp, we decided to use Q5® High-Fidelity DNA Polymerases, but others by OneTaq® 2X Master Mix. 2*UAS without template because the forward primer and reverse primer coμld combine each other and extend.<br> |
| - | The PCR systems set by protocol show below, we made 50μl reaction. | + | '''The PCR systems set by protocol show below, we made 50μl reaction.''' |
{{SUSTC-Image|wiki/images/7/7f/2014_SUSTC-Shenzhen_taq_PCR_protocol.png}} | {{SUSTC-Image|wiki/images/7/7f/2014_SUSTC-Shenzhen_taq_PCR_protocol.png}} | ||
{{SUSTC-Image|wiki/images/0/08/2014_SUSTC-Shenzhen_Q5_PCR_protocol.png}} | {{SUSTC-Image|wiki/images/0/08/2014_SUSTC-Shenzhen_Q5_PCR_protocol.png}} | ||
Revision as of 20:38, 15 October 2014
Parts building
design and building
Contents |
By Rifei Chen
For the linear backbone were so few and the backbone of BBa_J04450 was pSB1C3, we decide to cut it by XbaI and PstI restriction enzyme and recycle the about 2000bp vector as the backbone for biobricks. Of course, one advantage to make it as backbone is that we can recognize whether the gene we designed have inserted. Because if not, the bacterium will express red chromoprotein which one we unnecessary to pick up.
Week 4, Aug
2014/08/22
Transfect the backbone of biobricks
- Add 10μl ddwater to the plate, wait 10min, and absorb all the liquid into a PCR tube.
- Took a tube of supercompetent cells from -80℃ fridge, Immediately place the tube on ice, allow cell to thaw on ice, 10 min.
- Check the cell to see if they have thawed, gently flick the cells 1-2 times to evenly resuspend the cells.
- Mark the label with BBa_J04450.
- Add 2μl plasmid to the tube, shaking slightly.
- Incubate on ice for 30min.
- Heat shock, 42℃, 90s.
- Keep on ice immediately, 2 min.
- Add 200μl LB medium to each tube, shake with 37°C, 200rpm, 45min.
- Centrifuge, 4000rmp, 3min at room template.
- Discard most of the medium and reserve 50μl, resuspend by tips.
- Flam the SS-Spreader; put aside to cool; coated plates.
- Incubate the plates at 37°C, 16 hours.
2014/08/23
Shake the bacterium in tubes
- Add 3ml LB broth with ampicillin antibiotics into a tube.
- Flam the tweezers to red; put aside to cool.
- Pick up a tip which was sterilized; pick monoclone and throw the tip with monoclone into the tube directly.
- Shake with 37°C, 200rpm, and overnight.
Week 5, Aug
2014/08/24
Plasmid purification
- Pour 1.5ml of broth into a centrifuge tube, 11000 rpm, 2min.
- Repeat step1 again.
- Using TIANprep Mini Plasmid Kit, remove the medium completely, add 150μl P1 buffer, resuspend by tips, votex.
- Add 150μl P2 buffer, up and down gently 6~8 times; till it turn clear.
- Add 350μl P5, immediately invert up and down 10~12 times; till it clear and appear flocks. 11000rpm, 5 min.
- Transfer the supernatant into absorption column CP3, do not carry the precipitate. 11000rpm, 1 min. Discard the liquid waste.
- Add 600μl PW buffer; wait 3min; 11000rpm, 1min. Discard the liquid waste.
- Repeat step7 again.
- Centrifuge again at 11000 rpm, 2 min. Discard the liquid waste.
- Put the absorption column CP3 into a new centrifuge tube, open the cover and wait alcohol to spread completely.
- Add 70μl TB buffer into the absorption column center; wait 2min, 11000rpm, 2min.
- Re-suck up TB buffer which were centrifuged and add into the absorption column center again; wait 2min, 11000rpm, 2min.
- Measure the concentration of plasmid by Nanodrop2000. Mark in the tube.
2014/08/28
Design the biobricks
Week 2, Sep
2014/09/07
Design the primers
2014/09/11
Place an order of primers and completely synthesis
- Primers show:
SV40 promoter For: T TCTAGA G CGAACTGTGGAATGTGTG
SV40 promoter Rev: A GGATCC AT GAAGAC GACGAAAATGGATATACAAG
SV40 PolyA For: G AGATCT AT GAAGAC CCAACTTGTTTATTGCAGCTTA
SV40 PolyA Rev: CTGCAG CGGCCGC T ACTAGT ATCCATGCCGAGAGTGATGAA
pTRE-3G promoter For: T TCTAGA G TTTAAACTTTACTCCCTATC
pTRE-3G promoter Rev: A GGATCC AT GAAGAC GATTTACGAGGGTAGGAAGTG
TetOn For: T TCTAGA TGGGATCAAGACTGGACAAGA
TetOn Rev: CTGCAG CGGCCGC T ACTAGT A CCGAAGCCCAACCTTTCATA
CBh promoter For: T TCTAGA G TCTAGAGGTACCCGTTACA
CBh promoter Rev: A GGATCC AT GAAGAC GTCCAACCTGAAAAAAAGTGA
PB3 For: G AGATCT AT GAAGAC TGCTAGACTATAACAAGAA
PB3 Rev: CTGCAG CGGCCGC T ACTAGT A CGAAATTAACCCTCACTAA
PB5 For: T TCTAGA G ATTAGAAACTATTATTTAAC
PB5 Rev: A GGATCC AT GAAGAC GACACACATTCCACAGTTCG
- Completely synthesis show:
HBV1:
5'GATCAAGAAGACCTCACCAGGACCCCTGCTCGTGTTACAGGGTTTGGGTCTTCGATCTC 3'
HBV2:
5'GATCAAGAAGACCTCACCTCCGCAGTATGGATCGGCAGGTTTGGGTCTTCGATCTC 3'
HIV:
5'GATCAAGAAGACCTCACCTCTAGCAGTGGCGCCCGAACAGGGTTTGGGTCTTCGATCGA
5*UAS:
5'CGGAATTCGCGGCCGCTTCTAGAGCGGAGTACTGTCCTCCGAGCGGAGTACTGTCCTCCG
ACTCGAGCGGAGTACTGTCCTCCGATCGGAGTACTGTCCTCCGCGATTTCCGGAGTACTGTC
CTCCGTACTAGTAGCGGCCGCTGCAGAACCAATGCATTGG 3'
7*UAS:
5'CGGAATTCGCGGCCGCTTCTAGAGCGGAGTACTGTCCTCCGCGGAGTACTGTCCTCCGGA
ATTGCGATAGGTACCGAGTTACTAGACGGAGTACTGTCCTCCGAGCGGAGTACTGTCCTCCG
ACTCGAGCGGAGTACTGTCCTCCGATCGGAGTACTGTCCTCCGCGATTTCCGGAGTACTGTC
CTCCGTACTAGTAGCGGCCGCTGCAGAACCAATGCATTGG 3'
PolyA:
5'GAAGATCTTCGAAGACCTAGAGCTCGCTGATCAGCCTCGACTGTGCCTTCTAGTTGCCAG
CCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGCCACTCCCACTGT
CCTTTCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGTAGGTGTCATTCTATTCTGG
GGGGTGGGGTGGGGCAGGACAGCAAGGGGGAGGATTGGGAAGAGAATAGCAGGCATGCTG
GGGATACTAGTAGCGGCCGCTGCAGAACCAATGCATTGG 3'
NLS+NLS:
5'TCTGGAATTCGCGGCCGCTTCTAGAGCCAGATGGCCCCAAAGAAGAAGCGGAAGGTCGGT
ATCCACGGAGTCCCAGCAGCCACGTCTTCATGGATCCTATGAAGACTGAAAAGGCCGGCGGC
CACGAAAAAGGCCGGCCAGGCAAAAAAGAAAAAGTACTAGTAGCGGCCGCTGCAGTCCGGCA
AAAAAGGGC 3'
Week 3, Sep
2014/09/18
PCR TetOn, PB5, PB3, SV40/CBh/pTRE-3G promoter, PolyA1, 2*UAS
For TetOn and CBh were more than 700bp, we decided to use Q5® High-Fidelity DNA Polymerases, but others by OneTaq® 2X Master Mix. 2*UAS without template because the forward primer and reverse primer coμld combine each other and extend.
The PCR systems set by protocol show below, we made 50μl reaction.
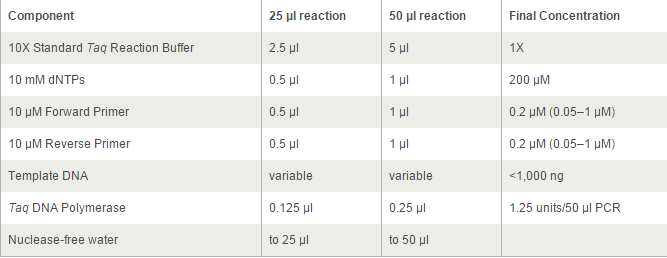
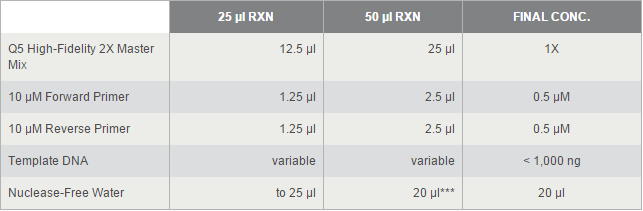 Thermocycling Conditions for a Routine PCR:
Thermocycling Conditions for a Routine PCR:
| Step | Temp (℃) | Time (s) |
|---|---|---|
| Initial denaturation | 98 | 90 |
| 30cycles | 98 | 30 |
| 48,52,55,58,60,63 | 30 | |
| 68 | 30 | |
| Final extension | 68 | 300 |
| Hold | 4 | ∞ |
Make a 1% Agarose gel:
| Component | Volume |
|---|---|
| 1×TAE buffer | 30ml |
| Agarose | 0.30g |
| Gene Green | 0.5ul |
Gel electrophoresis result:
PB3 and PB5 were both failed.
Purified others by TIANgel Midi Purification Kit. Use Nanodrop2000 to get each sample’s concentration. Digest TetOn, 2*UAS and backbone by XbaI and PstI overnight; CBh/SV40/pTRE-3G promoter by XbaI and BamHI; PolyA1 by BglII and PstI; purified.
 "
"