Team:Nagahama project
From 2014.igem.org
(→Modeling) |
(→Chemotaxis Assay) |
||
| Line 137: | Line 137: | ||
==Chemotaxis Assay== | ==Chemotaxis Assay== | ||
| + | ===Introduction=== | ||
| + | Chemotaxis by ''E.coli'' was assay by some methods. We assay the chemotaxis of ''E.coli'' against aspartate using two methods. One was swarming assay. We have used sawaming plate. swarming plate is medium that contained a low concentration by agar.'' E.coli'' can swim to aspartate on the swarming plate. The other was capillary assay. We have used capillary that including aspartate. We set capillary in chemotaxis medium including motile ''E.coli'' We have determined the number of ''E.coli'' attracted to capillary by colony formation on LB plate. | ||
===Swarmming Assay=== | ===Swarmming Assay=== | ||
===Capillary Assay=== | ===Capillary Assay=== | ||
Revision as of 17:11, 15 October 2014
| ||||||||||||||
Contents |
Our Project
Modeling
We consigned modeling of our project to
UT-Tokyo.They readily compiled with our requests. Their model is ideal.We really appreciate their jobs We wish a friendly relationship with UT-Tokyo.Thank you so much!! The detail of their jobs are lists below!!
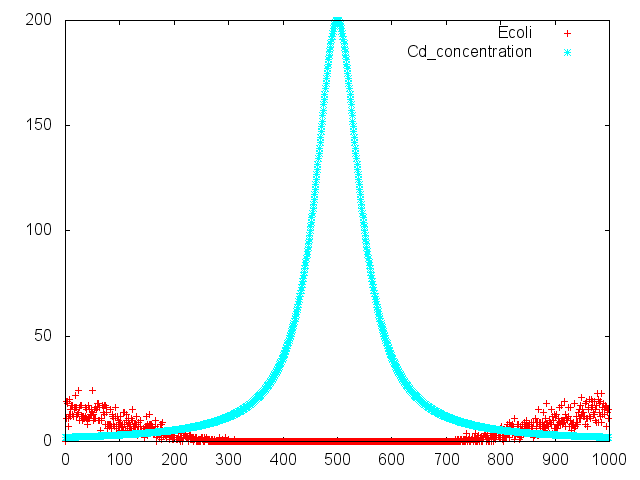
Fig.1 means random walk of E.coli on swarming plate.The vertical axis means arbitrary unit.The horizontal axis means spatial axis.
Fig.2 means positive chemotaxis by E.coli.Assuming that distill aspartate center of swarming plate. E.coli gather to distilled point.
Fig.3 means negativeChemotaxis by E.coli.Assuming that distill cadmium center of swarming plate. E.coli run away from distilled point.
Fig.4 means our project.
Assuming that distill cadmium (spatial axis 500). E.coli run away from distilled point (spatial axis 350~450 and 550~650). At that time E.coli`s cadmium promoter is operate. Asparatate is produced by E.coli.
Method
Medium
Aspartate synthesis
E.coli K12 transformed with CdP-R.B.S-AspA-d.Ter (BBa_K1342001) previous cultured with cadmium in LB medium (250μM) in 37℃ for 12hr at 120rpm. Adjust Cell mass (OD1.0) and therefor centrifuged 4000rpm for 20 min. Cell pellets ware activated in synthesis medium in 37℃ for 2hr at 120rpm/min.
SDS PAGE
・Preculture E.coli holding a plasmid containing a target gene or nomal E.coli.
・Measure OD600 0.6-1.0
・the expression of fusion protein by isopropylthio-β-D-galactoside (IPTG) and Cd2+ soln.
・Transfer a sample a 200 µl in a microcentrifuge tube
・Centrifuge at 13,000rpm for a minute at 4℃
・Discard supernatant quantitative
・Store pellet at -20 °C
・Thaw pellet and resupend in Sample Buffer (100 µL 1xSample Buffer per samples)
・Heat for 5 minutes at 98 °C
・Centrifuge at 13,000rpm for 10 minutes at 4℃
・Transfer supernatant to a new microcentrifuge tube
・Analyze samples by SDS-PAGE.(Use 20 µL per samples)
TLC assay
We analyzed L-aspartate by thin-layer chromatography (TLC). The same amount added Supernatant of synthesis medium to 7mg/mL 5-Dimethylamino naphthalene-1-sulfonyl Chloride (in Acetone) and incubated more than 30min. Two microliters of the sample was placed onto a TLC silica plate, and developed in a mixture of Ethyl acetate, pyridine, water, and acetic acid(162:21:11:6 V/V).
Chemotaxis Assay
Introduction
Chemotaxis by E.coli was assay by some methods. We assay the chemotaxis of E.coli against aspartate using two methods. One was swarming assay. We have used sawaming plate. swarming plate is medium that contained a low concentration by agar. E.coli can swim to aspartate on the swarming plate. The other was capillary assay. We have used capillary that including aspartate. We set capillary in chemotaxis medium including motile E.coli We have determined the number of E.coli attracted to capillary by colony formation on LB plate.
Swarmming Assay
Capillary Assay
Result & Discussion
TLC Assay

SDS PAGE
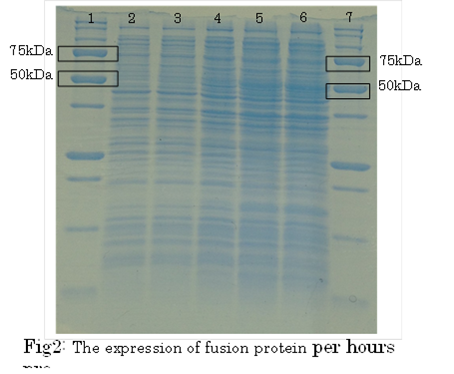 1. marker 2. 0hr 3. 0.5hr 4. 2hr 5. 6hr 6. 24hr 7. Marker
marker:Precision Plus Protein Standards (BIO-RAD)
1. marker 2. 0hr 3. 0.5hr 4. 2hr 5. 6hr 6. 24hr 7. Marker
marker:Precision Plus Protein Standards (BIO-RAD)
OD600=0.74
CdCl2 100μM/IPTG 1mM
 "
"