Team:Austin Texas/kit
From 2014.igem.org
Jordanmonk (Talk | contribs) m (→Experimental Design and Method) |
Jordanmonk (Talk | contribs) (→Experimental Design and Method) |
||
| Line 105: | Line 105: | ||
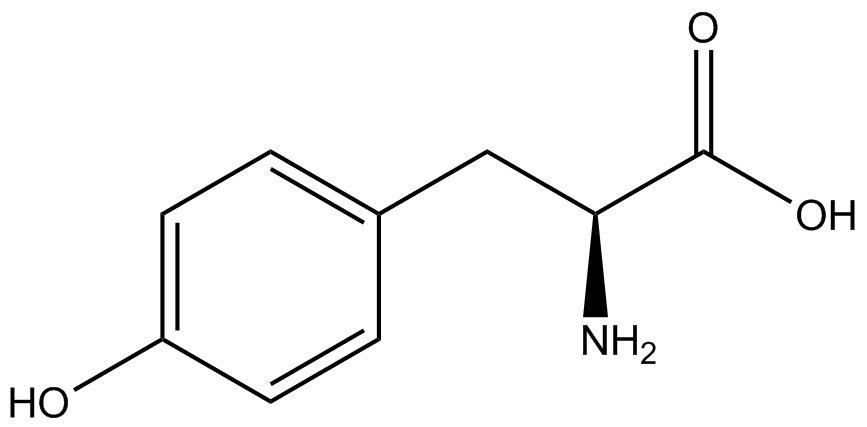
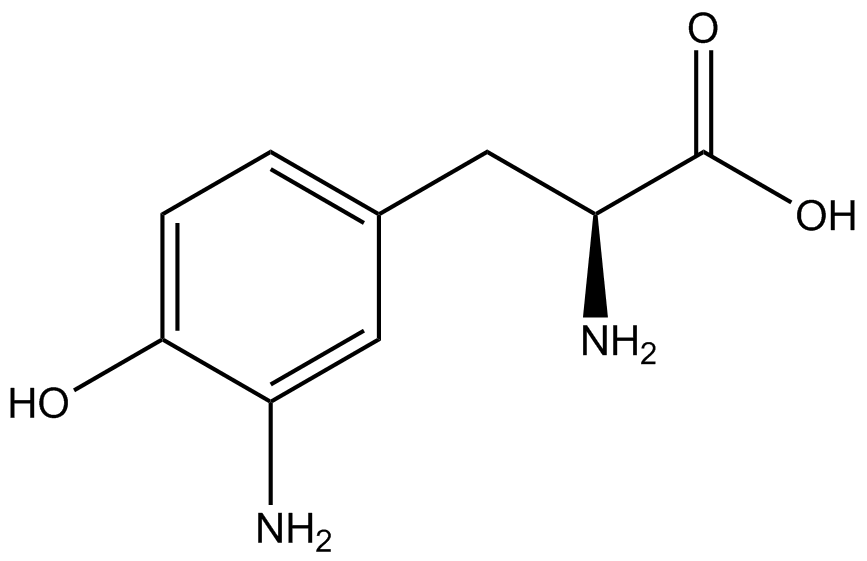
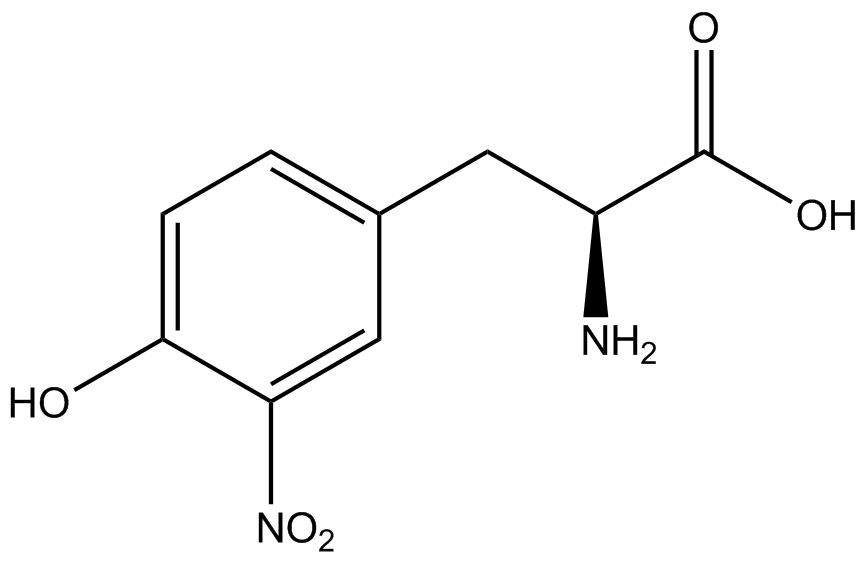
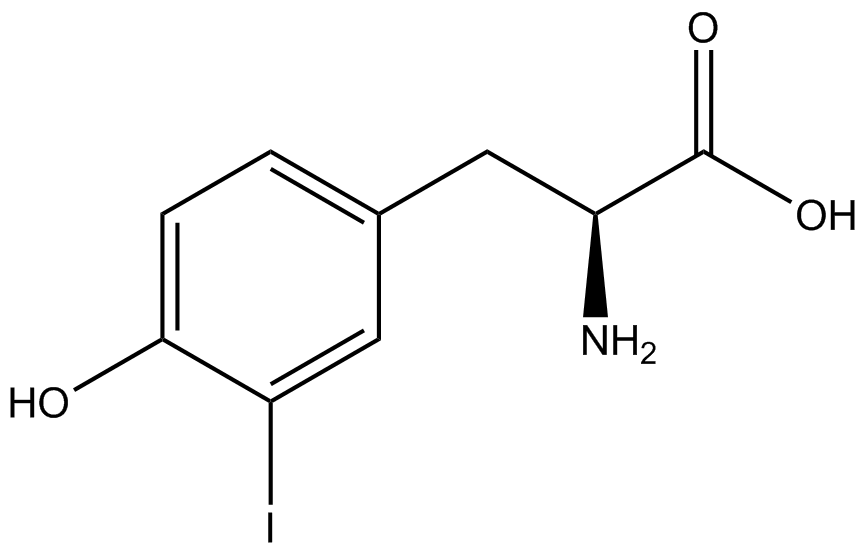
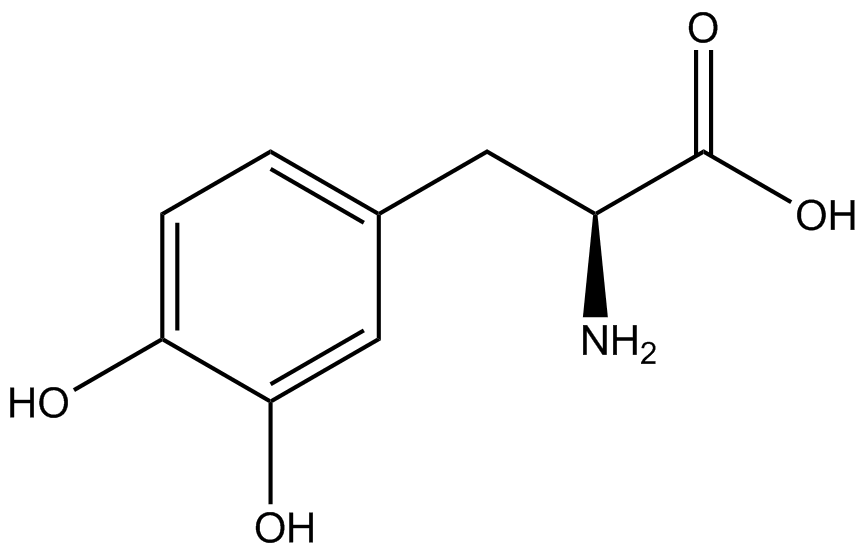
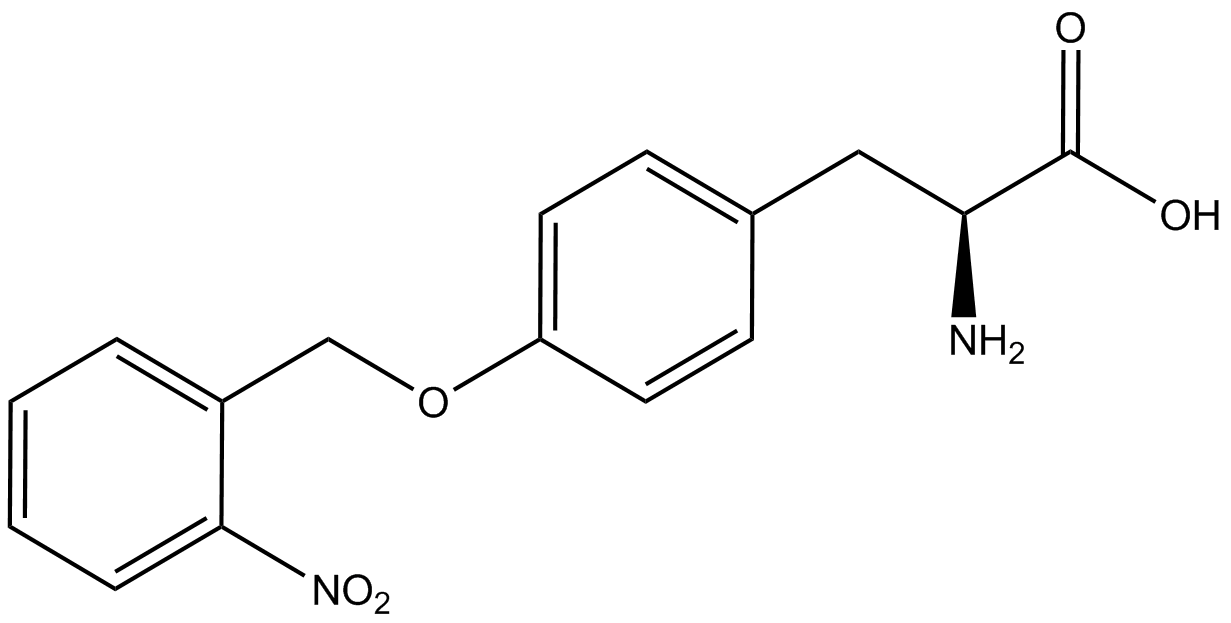
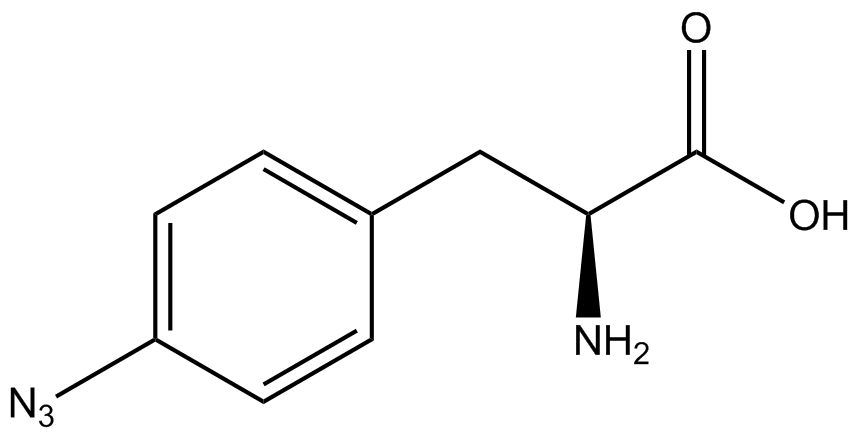
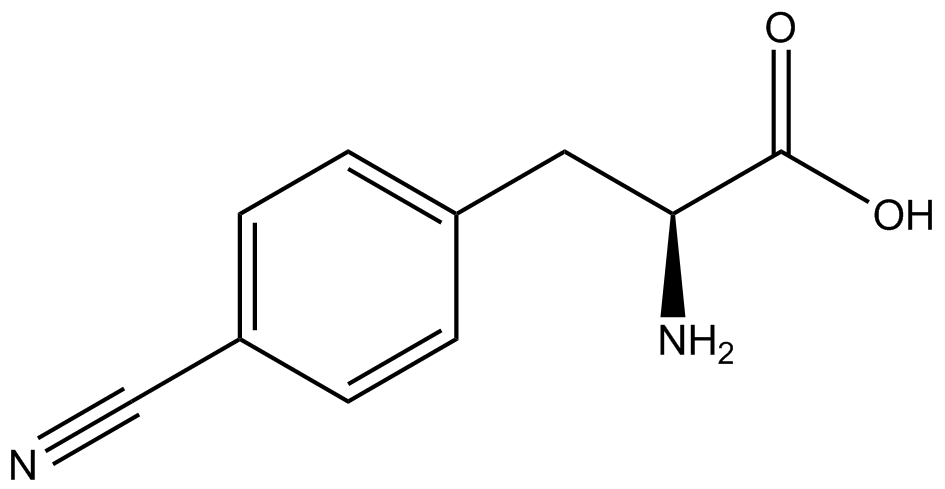
| - | [[File:Kit Plasmid Alignment 10-12-14.png|600px|thumb|right|Figure 2. Plasmids used in the ncAA kit.'''KATE, FIX the "pBIG" ALSO NO STOP CODON???''' | + | [[File:Kit Plasmid Alignment 10-12-14.png|600px|thumb|right|Figure 2. Plasmids used in the ncAA kit.'''KATE, FIX the "pBIG" to BLG ALSO NO STOP CODON???-Dennis''' ''Also, I would rotate the FRY plasmids 90º and BLG so the synthetase and tRNA sit top-middle and gent on the bottom-middle, remove the arrowhead on lacO and make it as narrow as the text, it's an operator, not a gene, and consider changing the name of BLG, the L was for lacI and that was moved to the FRY plasmids, right?-Jordan'' pBLG contains the specific synthetase/tRNA pair being tested. pFRYC and pFRY are the ncAA kit reporter plasmids. pFRYC is the control plasmid that yields GFP and RFP expression regardless of synthetase/tRNA pair. pFRY is a nearly identical plasmid with a single difference, the linker between the GFP and RFP sequences contains an amber codon in place of the tyrosine codon found in pFRYC. Thus, the level of GFP expression for pFRY is directly dependent on the efficiency and fidelity of the synthetase/tRNA pair being tested.]] |
| - | An ncAA synthetase/tRNA pair was cloned into pBLG and transformed into pFRYC amberless ''E. coli''. and pFRY amberless ''E. coli''. Other necessary control strains include RFP amberless ''E. coli'' (RFP control), sfGFP amberless ''E. coli'' (GFP control), amberless ''E. coli'' (cell background control), and LB media supplemented with ncAA | + | An ncAA synthetase/tRNA pair was cloned into pBLG and transformed into pFRYC amberless ''E. coli''. and pFRY amberless ''E. coli''. Other necessary control strains include RFP amberless ''E. coli'' (RFP control), sfGFP amberless ''E. coli'' (GFP control), amberless ''E. coli'' (cell background control), and LB media supplemented with ncAA (media background control). An overnight culture of each strain was grown in LB with the appropriate antibiotics at 37ºC and 225rpm. 10 mL of media with the appropriate antibiotics was inoculated with 100 µL of overnight culture and allowed to grow in the same conditions until the culture density was ~0.2-0.3 OD, or ~3 hours. The 10 mL culture was split between 4 different sterile test-tubes, 2 mL of culture per tube. The conditions of test tubes A through D were as follows: A (-IPTG,-ncAA), B (-IPTG,+ncAA), C (+IPTG, -ncAA), and D (+IPTG, +ncAA). IPTG stock solution was made at 1000X concentration ('''why is this here?''') and the ncAA was added to yield a concentration of 1 mM. Sterile deionized water was added in the place of ncAA and IPTG as a control ('''?'''). Once the controls, IPTG, and the ncAA were added appropriately, the cultures were allowed to grow to ~0.5 OD. 70 µL of each culture condition and control culture was added to a separate wells in a transparent 96-well plate for fluorescence and OD readings in a microplate reader. |
Revision as of 09:02, 14 October 2014
|
 "
"