Team:Austin Texas/photocage
From 2014.igem.org
(→Background) |
|||
| Line 93: | Line 93: | ||
=Background= | =Background= | ||
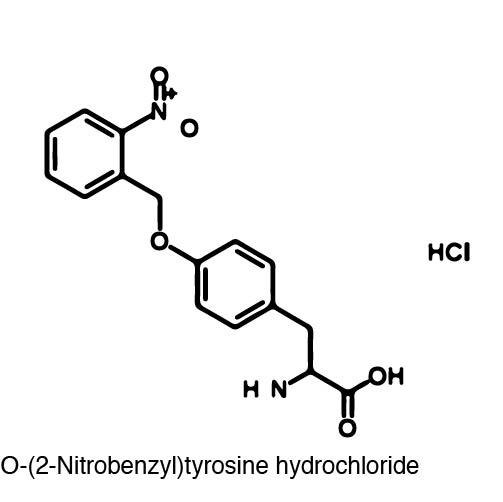
| - | + | Tyrosine residue 639 (Y639) was specifically targeted because it lies on a crucial position on the O-helix and has been proved to be essential for polymerization. (REFERENCE) The Y639 residue in the O-helix is responsible for two major roles. First, this tyrosine residue discriminates between deoxyribose and ribose substrates. Second, Y639 is responsible for moving newly synthesized RNA out of the catalytic site and preparing for the next NTP to be inserted. These functions of the O-helix were shown to be essential through mutational analysis. Because the loss of this tyrosine residue in the active site leads to a non-functional polymerase, Y639 proved to be a good candidate for incorporating a photocaged amino acid (Reference; You are referring to a specific result). | |
| - | + | Incorporation of ONBY at position 639 halts the activity of the T7 polymerase because of the bulky nature of ONBY (REFERENCE). However, the bulky ONB group is able to be removed through irradiation with 365 nm light. The wavelength of light used to "decage" the amino acid proved to be another advantage because 365 nm light is not toxic to the cell (accurate?). Once the ONB group is removed, a normal tyrosine residue is left which restores RNA polymerase activity. | |
| - | In order to incorporate the | + | In order to incorporate the ncAA into amberless E.coli (which is described '''[here]'''), a ''Methanocaldoccus jannaschii'' tyrosyl-tRNA synthetase/tRNA pair was previously mutated to selectively charge and incorporate ONBY. Six residues (Tyr 32, Leu 65, Phe 108, Gln 109, Asp 158, and Leu 162) on the original synthetase were randomized and the library was selected for its ability to charge ONBY while discriminating against other canonical amino acids. The resulting mutant ONBY synthetase contained five mutations (reference). The Asp 158→Ser 158 and Tyr 32→Gly 32 mutations are believed to result in the loss of hydrogen bonds with the natural substrate, which would disfavor binding to tyrosine, while the Tyr 32→Gly 32 and Leu 65→Gly 65 mutations are believed to increase the size of the substrate-binding pocket to accommodate the bulky o-nitrobenzyl group. The fifth mutation is ?? ARE THESE MUTATIONS CORRECT? Y32G is listed twice, which is fine, but then we should list the other mutations and what their roles are or that their roles are unknown. |
Revision as of 17:55, 12 October 2014
|
 "
"