Team:Austin Texas/photocage
From 2014.igem.org
(→Introduction) |
(→Introduction) |
||
| Line 78: | Line 78: | ||
=Introduction= | =Introduction= | ||
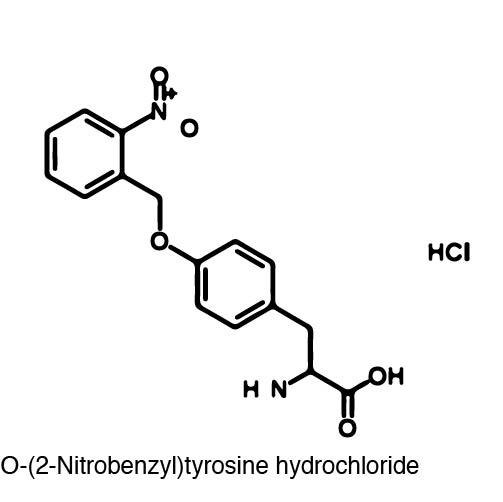
| - | [[Image:orthonitrobenzyl_tyrosine_structure.jpg |left| 250px ]] | + | [[Image:orthonitrobenzyl_tyrosine_structure.jpg |left| 250px|thumb| The ONBY ncAA used for our photocaging project. When exposed to 365 nm light, the ONB group is released, resulting in a normal tyrosine amino acid.]] |
| + | |||
| + | |||
Using non-canonical amino acid incorporation, we recreated a light-activatable T7 polymerase (RNAP) for the spatio-temporal control of protein expression. The light-activatable T7 RNAP was created by mutating a Tyrosine codon at position 639 of the O-helix (figure??? of helix?), which is a domain crucial for polymerization of RNA during transcription. For this project, Y639 was mutated to an amber stop codon and we used ortho-nitrobenzyl tyrosine (ONBY), which is a "photocaged" ncAA. Thus, if our synthetase/tRNA pair works, position 639 should contain ONBY in place of Y. This work is essentially a recapitulation of earlier work done by [reference authors/paper]. | Using non-canonical amino acid incorporation, we recreated a light-activatable T7 polymerase (RNAP) for the spatio-temporal control of protein expression. The light-activatable T7 RNAP was created by mutating a Tyrosine codon at position 639 of the O-helix (figure??? of helix?), which is a domain crucial for polymerization of RNA during transcription. For this project, Y639 was mutated to an amber stop codon and we used ortho-nitrobenzyl tyrosine (ONBY), which is a "photocaged" ncAA. Thus, if our synthetase/tRNA pair works, position 639 should contain ONBY in place of Y. This work is essentially a recapitulation of earlier work done by [reference authors/paper]. | ||
Revision as of 17:41, 12 October 2014
|
 "
"