Team:Austin Texas/photocage
From 2014.igem.org
Nathanshin (Talk | contribs) |
(→Introduction) |
||
| Line 78: | Line 78: | ||
=Introduction= | =Introduction= | ||
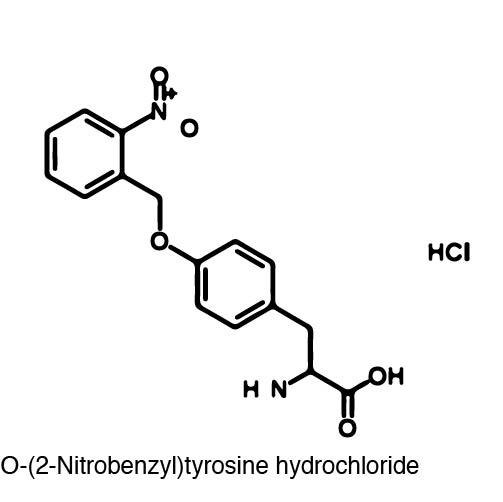
| - | + | [[Image:orthonitrobenzyl_tyrosine_structure.jpg |left| 250px ]] | |
| - | + | Using non-canonical amino acid incorporation, we recreated a light-activatable T7 polymerase (RNAP) for the spatio-temporal control of protein expression. The light-activatable T7 RNAP was created by mutating a Tyrosine codon at position 639 of the O-helix (figure??? of helix?), which is a domain crucial for polymerization of RNA during transcription. For this project, Y639 was mutated to an amber stop codon and we used ortho-nitrobenzyl tyrosine (ONBY), which is a "photocaged" ncAA. Thus, if our synthetase/tRNA pair works, position 639 should contain ONBY in place of Y. This work is essentially a recapitulation of earlier work done by [reference authors/paper]. | |
| + | |||
| + | |||
| + | |||
| + | When the photocaged ONBY is incorporated at position 639, RNAP activity is halted. The polymerase can become activated only upon de-caging of the ONB group from the ONBY. This is accomplished by exposing the cells to 365 nm light. When exposed to 365 nm light, the ONB group is released, resulting in position 639 now containing a normal tyrosine amino acid. T7 RNAP was selected because of its orthogonal nature, which allows us to selectively induce the expression of specific genes that are preceded by the T7 RNAP promoter. Because T7 promoters are not natively found in E.Coli, a gene downstream of a T7 promoter may be exclusively expressed through the introduction of 365 nm light. | ||
=Background= | =Background= | ||
Revision as of 17:40, 12 October 2014
|
 "
"