Team:Aachen/Project/2D Biosensor
From 2014.igem.org
(→Principle of Operation) |
(→Principle of Operation) |
||
| Line 11: | Line 11: | ||
''Add: Flow chart of chip measurement'' | ''Add: Flow chart of chip measurement'' | ||
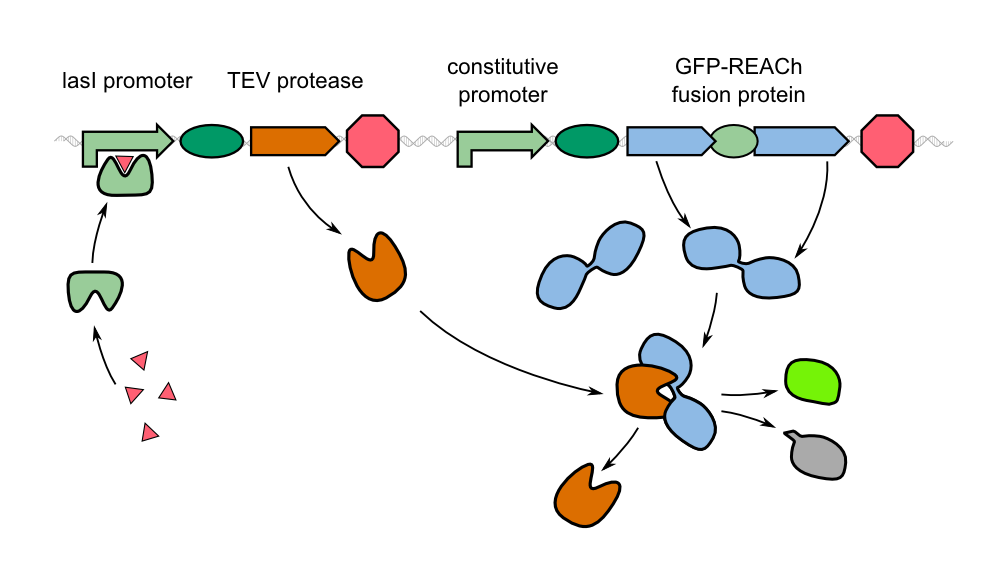
| - | To detect ''P. aeruginosa'' cells, the chip is placed on a hard surface that is potentially contaminated with the pathogen. Subsequently, the chip is introduced into our measurement device ''WatsOn'' where it is incubated at 37 °C. After a short time, the chip is illuminated with blue light. ''WatsOn'' takes a picture of the chip and the software ''Measurarty'' analyzes any fluorescent signal. Depending on the intensity of the signal and the size of the spot, ''Cellock Holmes'' can '''calculate concentration and distribution of ''P. aeruginosa'' ''' on the sampled surface. | + | To detect ''P. aeruginosa'' cells, the chip is placed on a hard surface that is potentially contaminated with the pathogen. Subsequently, the chip is introduced into our measurement device [https://2014.igem.org/Team:Aachen/Project/Measurement_Device ''WatsOn''] where it is incubated at 37 °C. After a short time, the chip is illuminated with blue light. ''WatsOn'' takes a picture of the chip and the software ''Measurarty'' analyzes any fluorescent signal. Depending on the intensity of the signal and the size of the spot, ''Cellock Holmes'' can '''calculate concentration and distribution of ''P. aeruginosa'' ''' on the sampled surface. |
Revision as of 15:07, 10 October 2014
|
|
 "
"