Team:Oxford/biosensor deterministic equations
From 2014.igem.org
(Difference between revisions)
Olivervince (Talk | contribs) |
Olivervince (Talk | contribs) |
||
| Line 277: | Line 277: | ||
<div class="pink_news_block"> | <div class="pink_news_block"> | ||
<h1>Modelling the activations and repressions in our genetic circuit</h1> | <h1>Modelling the activations and repressions in our genetic circuit</h1> | ||
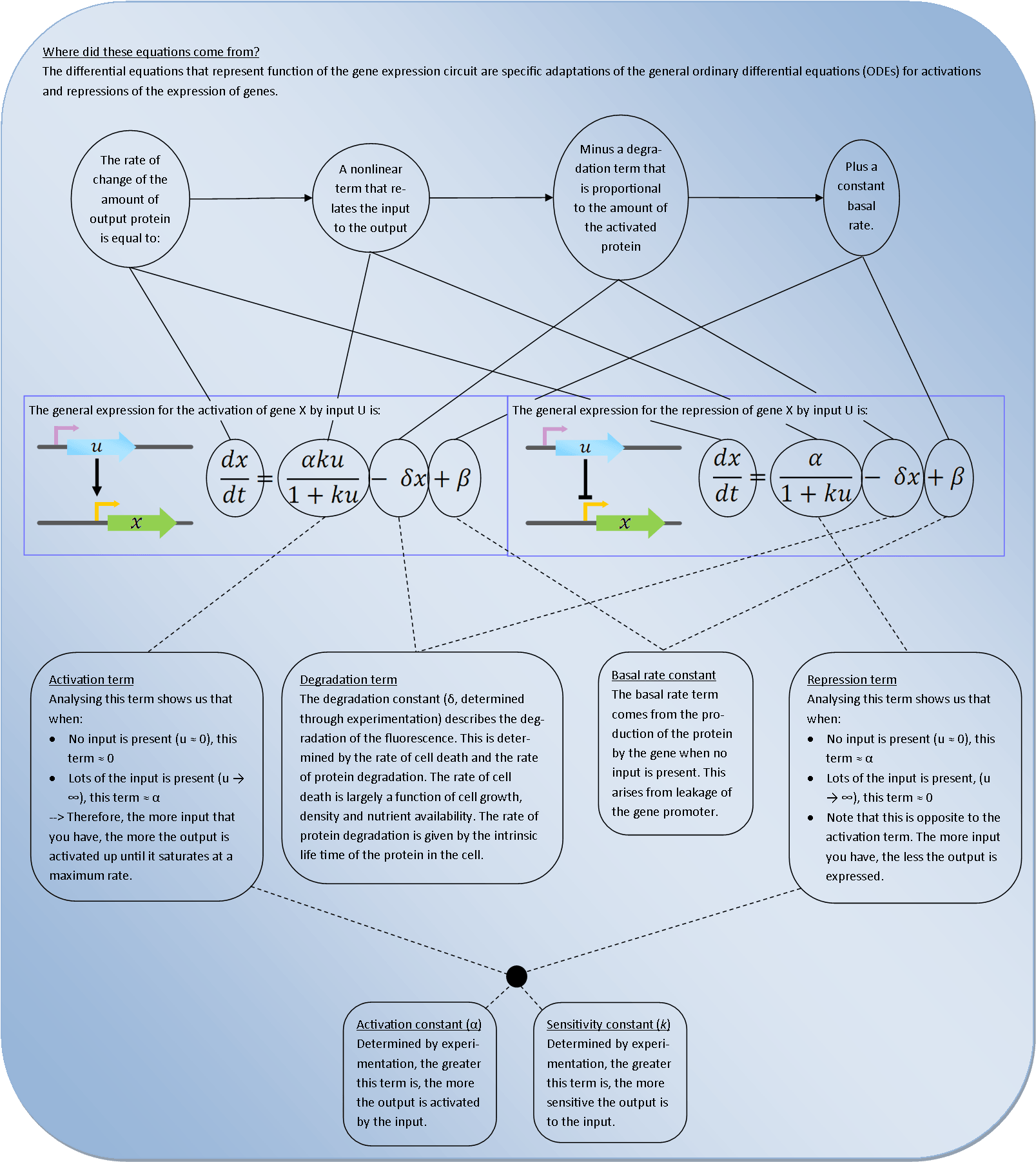
| - | To deterministically model each activation and repression of promoters by proteins, we used first order ordinary differential equations (ODEs). | + | To deterministically model each activation and repression of promoters by proteins, we used first order ordinary differential equations (ODEs). The graphic below explains where all of the terms come from and what effect they have on the system. |
<br><br> | <br><br> | ||
| - | This explanation should enable people of any academic discipline to gain | + | This explanation should enable people of any academic discipline to gain an understanding of how to begin simulating gene circuits and predicting the reponse of their systems. |
</div> | </div> | ||
Revision as of 15:06, 7 October 2014
 "
"