Team:KAIT Japan/Protocol
From 2014.igem.org
(Difference between revisions)
| (43 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{|style= width="100%" align="center" | {|style= width="100%" align="center" | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New home t.jpg|link=Team:KAIT JAPAN]] |
[[Team:KAIT JAPAN|Home]] | [[Team:KAIT JAPAN|Home]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New Team1.jpg|link=Team:KAIT_Japan/Team]] |
[[Team:KAIT_Japan/Team|Team]] | [[Team:KAIT_Japan/Team|Team]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New 考える人.jpg|link=Team:KAIT_Japan/Project]] |
[[Team:KAIT_Japan/Project|Project]] | [[Team:KAIT_Japan/Project|Project]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New 歯車.jpg|link=Team:KAIT_Japan/Parts]] |
[[Team:KAIT_Japan/Parts|Parts]] | [[Team:KAIT_Japan/Parts|Parts]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New 111111111111111.jpg|link=Team:KAIT_Japan/Protocol]] |
[[Team:KAIT_Japan/Protocol|Protocol]] | [[Team:KAIT_Japan/Protocol|Protocol]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:Ff.jpg|link=Team:KAIT_Japan/Notebook]] |
[[Team:KAIT_Japan/Notebook|Notebook]] | [[Team:KAIT_Japan/Notebook|Notebook]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New 結果.jpg|link=Team:KAIT_Japan/Results]] |
[[Team:KAIT_Japan/Results|Results]] | [[Team:KAIT_Japan/Results|Results]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New new 危険.jpg|link=Team:KAIT_Japan/Safety]] |
[[Team:KAIT_Japan/Safety|Safety]] | [[Team:KAIT_Japan/Safety|Safety]] | ||
|} | |} | ||
| | | | ||
| - | {|style="background:# | + | {|style="background:#FFFFFF; margin:0.1em" |
| - | !align="center"|[[File: | + | !align="center"|[[File:New new ひと.jpg|link=Team:KAIT_Japan/Human_Practice]] |
[[Team:KAIT_Japan/Human_Practice|Human Practice]] | [[Team:KAIT_Japan/Human_Practice|Human Practice]] | ||
|} | |} | ||
| | | | ||
|} | |} | ||
| - | {| style="width:98.5%; margin-left:0.2em; border-color:#00BFFF; border-style:solid; border-collapse:collapse; background-color:# | + | {| style="width:98.5%; margin-left:0.2em; border-color:#00BFFF; border-style:solid; border-collapse:collapse; background-color:#ffffe0; text-align:left" |
| - | | style="width: | + | | style="width:98.5% style="vertical-align:top" | |
| + | |||
| + | |||
<H1><font size="7">Protocol</font></H1> | <H1><font size="7">Protocol</font></H1> | ||
| Line 153: | Line 155: | ||
:23) We stored low temperature | :23) We stored low temperature | ||
</p> | </p> | ||
| + | |||
<br> | <br> | ||
<br> | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
<font size="5">2:PCR</font> | <font size="5">2:PCR</font> | ||
<p> | <p> | ||
| - | :'''・We performed PCR to confirm whether DNA which | + | :'''・We performed PCR to confirm whether DNA which we need.''' |
</p> | </p> | ||
| Line 166: | Line 172: | ||
<p> | <p> | ||
| - | :2) | + | :2)Made PCR preparation liquid {buffer×10:2.5ul,dNTP:2ul,PrimerF:1ul,PrimerR:1ul,Taq Polumerase:0.1ul,D<sub>2</sub>W:17.39ul /1 microcentrifuge tube(0.2ml)} |
</p> | </p> | ||
| Line 172: | Line 178: | ||
:3)Added sample(there are Plasmid made with miniprep)to the microcentrifuge tube and do PCR(The PCR conditions are as follows). | :3)Added sample(there are Plasmid made with miniprep)to the microcentrifuge tube and do PCR(The PCR conditions are as follows). | ||
</p> | </p> | ||
| + | |||
| + | <p> | ||
| + | :4)We did Electrophoresis to confirm Objective band. | ||
| + | </p> | ||
| + | |||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
| - | + | <br> | |
| - | + | <br> | |
| - | + | ||
<p> | <p> | ||
| - | : | + | <font size="5">3:Restriction enzyme processing</font> |
</p> | </p> | ||
| - | + | <p> | |
| + | :'''・We did restriction enzyme processing to plasmid to incorporate inserts in a plasmid vector.''' | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
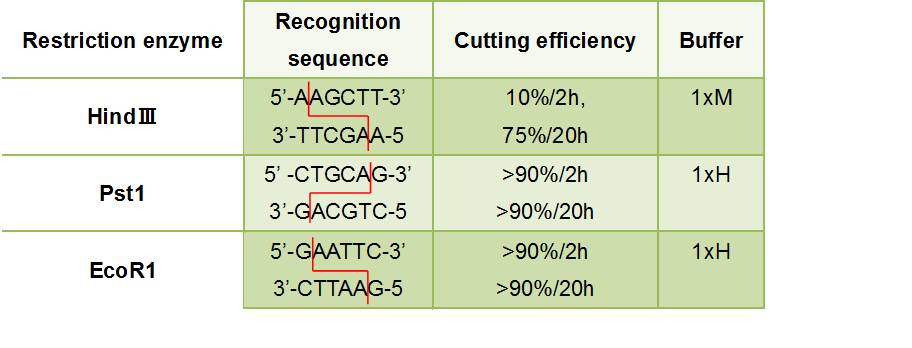
| + | :::Figure②:Used Restriction enzyme<br> | ||
| + | :[[File:かか.jpg]] | ||
| + | </p> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <p> | ||
| + | :<font size="4">'''HlyA'''</font>・・・We removed the stop codon<br> | ||
| + | ::F chain ・・・This has EcoR1 recognition sequence.We added <font color="#FFA500">CG</font> and <font color="#FF0000">A</font> to this to prevent flame out.<br> | ||
| + | :::::・5'-<font color="#FFA500">CG</font>GAATTC<font color="#FF0000">A</font>TTAGCCTATGGAAGTCAGGG-3'<br> | ||
| + | :::::((Tm before adding a restriction enzyme site =60℃<br> | ||
| + | :::::((GC before adding a restriction enzyme site =50.0% | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | ::R chain ・・・This has HindⅢ recognition sequence.We added <font color="#FFA500">CCC</font> to this.<br> | ||
| + | :::::・5'-<font color="#FFA500">CCC</font>AAGCTTTGCTGATGTGGTCAGGGTTA-3'<br> | ||
| + | :::::((Tm before adding a restriction enzyme site =60℃<br> | ||
| + | :::::((GC before adding a restriction enzyme site =50.0% | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :<font size="4">'''GFP'''</font><br> | ||
| + | ::F chain ・・・This has HindⅢ recognition sequence.We added <font color="#FFA500">CCC</font> and <font color="#FF0000">A</font> to this to prevent flame out.<br> | ||
| + | :::::・5'-<font color="#FFA500">CCC</font>AAGCTT<font color="#FF0000">A</font>ATGCGTAAAGGAGAAGAACT-3'<br> | ||
| + | :::::((Tm before adding a restriction enzyme site =56℃<br> | ||
| + | :::::((GC before adding a restriction enzyme site =40.0% | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | ::R chain ・・・This has Pst1 recognition sequence.We added <font color="#FFA500">AA</font> to this.<br> | ||
| + | :::::・5'-<font color="#FFA500">AA</font>CTGCAGTTATTATTTGTATAGTTCATCC-3'<br> | ||
| + | :::::((Tm before adding a restriction enzyme site =54℃<br> | ||
| + | :::::((GC before adding a restriction enzyme site =22.7% | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
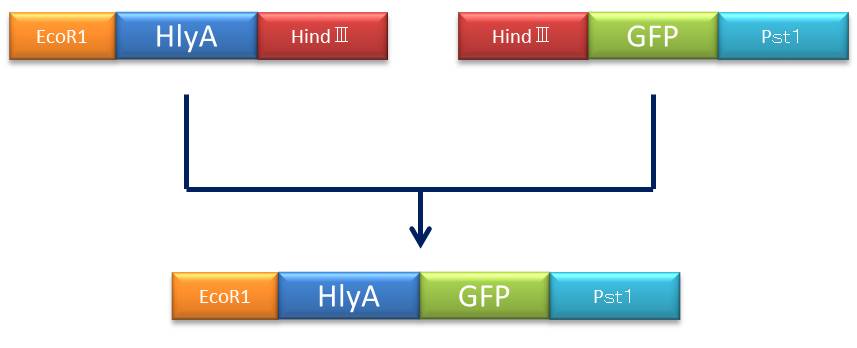
| + | :[[File:プラン1.jpg]] | ||
| + | <br> | ||
| + | ::::::::::::<font size="4">Figure③:Rough plan</font> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <p> | ||
| + | :1)We set a heat block(37℃). | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :2)prepare one microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :3)added 1ul restriction enzyme buffer(×10) to microtube | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :4)added 1ul DNA sample liquid to the microtube | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :5)added 1ul each restriction enzyme to the microtube | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :6)added D<sub>2</sub>W until it became 10ul | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :7)set microtube to a heat block (37℃) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :8)left the microtue more than one hour to push forward a reaction | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | <font size="5">4:Ligation</font> | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :1)set the temperature of the heat block. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :2)prepared one microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :3)added 2ul PCR product (insert DNA) to the microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :4)added 1ul Plasmid vector (did with restriction enzyme) to the microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :5)added 5ul DNA ligase to the microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :6)added 5ul Sterilization water to the microtube and closed the cap | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :7)set the microtube to heat block. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :8)put it for several hours. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :9)after 8),saved it in -20℃. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | <p> | ||
| + | <font size="5">5:Transformation</font> | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :1)We set a water bus(42℃) and a heat block(37℃). | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :2)added 280ul LB media to empty microtube and set to heat block. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :3)added plasmid(1ul or 2ul) to microtube which has 10ul competent cell. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :4)Did tap. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :5)left this microtube on ice for 15 minutes. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :6)did heat shock with a water bus for 45 seconds. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :7)brought it to ice quickly and left on ice for 2 min. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :8)added 250ul LB media to this. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :9)cultured it at 37 ℃ for one hour | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :10)applied to LB agar nutrient medium about 20ul arabinose (200 mg/ml) and 20ul ampicillin (100 mg/ml) While I culture it. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :11)After 9),we added 125ul Bacteria liquid (we cultured in 9)) to LB agar nutrient medium (we made in 10)). | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | <p> | ||
| + | <font size="5">6:DNA purification</font> | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :1)From the gel which did electrophoresis, I cut and bring down an objective DNA band. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :2)moved the gel which I cut and brought down to microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :3)added 500ul buffer QG to the microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :4)incubated it for 10min.(50℃) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :5)moved liquid of 4) to QIA quick colum. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :6)Centrifugal separation(10000rpm, 2 min) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :7)discarded the liquid which collected at the bottom of QIA quick colum. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :8)Centrifugal separation(10000rpm, 1 min) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :9)added 750ul buffer PE to the QIA quick colum and put this about 2 min. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :10)Centrifugal separation(10000rpm, 2 min) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :11)repeated 9)~10) 2~3 times. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :12)discarded the liquid which collected at the bottom of QIA quick colum. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :13)Centrifugal separation(10000rpm, 4 min) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :14)discarded the liquid which collected at the bottom of QIA quick colum. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :15)put the QIA quick colum which disintegrated on the new microtube. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :16)added 50ul buffer EB to the QIA quick colum and put about 5 min. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :17)Centrifugal separation(10000rpm, 2 min) | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | :18)Objective DNA is in the bottom of the microtube. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
</font> | </font> | ||
Latest revision as of 04:47, 16 October 2014
|
|
|
|
|
|
|
|
|
|
Protocol1:miniprep
2:PCR
3:Restriction enzyme processing
4:Ligation
5:Transformation
6:DNA purification
|
 "
"