Team:UChicago/Notebook
From 2014.igem.org
| (48 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:UChicago/NavBar}} | {{Team:UChicago/NavBar}} | ||
| - | + | <html> | |
| + | <style> | ||
| - | + | #notebookpage { | |
| - | + | } | |
| - | == | + | </style> |
| + | <div id="notebookpage" class="maindiv"></html> | ||
| + | <h1>Lab Notebook</h1> | ||
| + | See [[Media:UChicago_LabNotebookSummary.pdf]] for the original document. | ||
| - | + | ==Week 1 6/16 - 6/20== | |
| - | + | ===Monday=== | |
| + | First day of lab! We plated some DH5-alpha competent cells in preparation for gDNA extraction for our PCRs and future transformations. One person showed up (Kevin yayyyyy), then left. | ||
| - | + | [[File:UChicago_kevin.jpg|200px]] | |
| - | + | ===Tuesday=== | |
| - | + | We attempted to make LB plates. We had fun watching it boiling over since we left it in the hot plate too long and it spilled everywhere. We did clean-up time afterwards! In the future, we will remember to to heat for only 1 min to dissolve everything except agar. Agar will dissolve during autoclave because science is cool. So exciting! :D We then remade the LB for funsies :D We did a few other things to start things up like making SOB, CCMB80 competent cell buffer, and antibiotic plates. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Wednesday=== | |
| - | + | MOAR BUFFERS! Qiagen buffers are expensive so we spent the day making bootleg Qiagen miniprep buffers (sorry Qiagen, you had [[Team:UChicago/Donate|your chance]]). Then, we contemplated life for the rest of the day. *take depressing pictures here* | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | *take pictures of buffers* | |
| - | * | + | |
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Thursday=== | |
| - | + | Made competent cells. Stuck hands in liquid nitrogen. Burnt our hands off (no pics because you need hands to use a camera, duh). EVEN MOAR BUFFERS | |
| - | + | We also went to a biotech company exhibit and grabbed a lot of free samples! (and food of course) | |
| - | + | We also extracted gDNA from DH5a to PCR out our mutator genes. We also learned not to use the free 1.5mL centrifuge tubes from last year, and to not smell our microcentrifuge anymore. | |
| + | Stock strains MG1655 and delta-tyrR came in from the Yale E Coli Stock Center. Yay! So exciting! Kevin was so excited that he burnt his hair trying to plate them. XD | ||
| - | + | We made competent cells (MY CRYSTAL BALL SAYS: THEY WILL FAIL). Kevin BS’d the gDNA and got some somehow but also made the centrifuge stink like chloroform/phenol. Fun XD | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:ipredictfailure.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | This company (name concealed) gave us some cool centrifuge tubes last year that warp when we spin them down. XD So we’re buying new ones. $$$ They’re wrong- money DOES grows on trees! AND we have lots of trees around here! Even better! We ran overnight PCRs, one with more and one with less cycles. | |
| - | + | [[File:uchicago_week_1_gel.jpg]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Friday=== | |
| - | + | PCR-ed out 9/12 mutators. The lengths of almost all of them were verified to be correct!!! So much happy!!! Yes!!!! Yay!!!! The PCR with less cycles (bottom half of the gel) worked better so we’ll stick with that in the future. We also digested backbones that have the RBS we will use and ligate to all the mutators. What a good day! | |
| - | + | ||
| - | + | ||
| - | == | + | ==Week 2 6/23 - 6/27== |
| - | + | ===Monday=== | |
| - | + | We tested our competent cells using the transformation efficiency kit. They didn’t work XD But that’s okay, we like making competent cells. It’s strangely exciting to pipette things lots of times :D | |
| - | + | ||
| - | + | ||
| - | + | Also, we fragment isolated the mutations and then did the restriction digests on the PCR products. The concentrations of the genes are fairly alright, about 20-50 ng/microL. We ran a gel of AroF to verify PCR of the gene, but unfortunately we did not get a band XD. That’s okay, life goes on even if your gels fail!!! XD IT’S summer time! Rainbows! | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | We gel extracted the mutators that were digested last week. Fun. Got high contamination. But contamination is good because what doesn’t kill your experiments makes them stronger right ? Yes! Concentrations are listed below XD | |
| - | + | ||
| - | + | {|class="wikitable" | |
| - | + | |+Mutator Concentrations | |
| - | + | |- | |
| - | + | !Sample||Conc (ng/microL)||260/280||260/230 | |
| + | |- | ||
| + | |ParC||30.2||1.78||0.25 | ||
| + | |- | ||
| + | |DinB||55.8||1.70||0.42 | ||
| + | |- | ||
| + | |MutS||21.3||1.86||0.18 | ||
| + | |- | ||
| + | |Dam||37.0||1.68||0.18 | ||
| + | |- | ||
| + | |MutT||29.9||1.61||0.09 | ||
| + | |- | ||
| + | |Umu’C||24.5||1.80||0.05 | ||
| + | |- | ||
| + | |EmrR||36.8||1.71||0.08 | ||
| + | |- | ||
| + | |DnaE||31.1||1.53||0.32 | ||
| + | |} | ||
| - | + | ===Tuesday=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | NOTHING WORKED :)))))!!!!!!!! We drank our worries away XD XD XD. We drunkenly made glycerol stocks of the two strains from Yale that came last Thursday. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | *hic* guuysh guysh we should *burrrrp* totally ligate a rBs to a vecter *hi*c | |
| - | * | + | Alcohol is poison. (Delicious, delicious poison. And we are Mithridates.) |
| - | * | + | |
| - | * | + | |
| - | + | ||
| - | + | ||
| + | Unfortunately, our cells weren’t competent when we tested them again :( We went over the protocol and made sure we’ll have everything down for next time XD. Or rather, fortunately! We get to make more! XD Soooo stoked! | ||
| + | We fragment isolated and gel purified our promoter plasmids. | ||
| + | For our project planning, we met with our faculty advisors Mike and Jennifer. The meeting was super, super, super productive and educational! We could consider inserting a polylinker into our plasmids to reduce a lot of steps XD Polylinkers are fun! | ||
| + | |||
| + | ===Wednesday=== | ||
| + | |||
| + | We purified the 8 mutators that we PCR-ed out. As for AroF and the three mutators that didn’t work, we tried PCR with Phusion, but unfortunately it didn’t work again. Yay! | ||
| + | After digesting our RBS and ligating it with our previous cut promoter plasmids, we also did 3A assembly in parallel for RBS and promoter. Yay! | ||
| + | More amp and tet plates were made! Yayyy! | ||
| + | *picture of plates” | ||
| + | |||
| + | |||
| + | ===Thursday=== | ||
| + | We had issues with SOC contamination today - We had to make a new batch for more transformations. We also tried transformation with the efficiency kit again, making sure we were following each step as it was written on our protocol. | ||
| + | Friday | ||
| + | Transformation of our 3A assembly colonies didn’t work, but the transformation of mcherry plasmids worked! This means that there’s nothing wrong with our transformation protocol. We will look into our digestion and ligation steps. We <3 troubleshooting! All part of the scientific process XD! | ||
| + | Here’s a joke XD: | ||
| + | What do you call it when McDonald sells cherries? | ||
| + | mCherry! HAHAHA XD! | ||
| + | |||
| + | We made more chloramphenicol plates again! Yay! | ||
| + | |||
| + | ==Week 3 6/30 - 7/4== | ||
| + | |||
| + | ===Monday=== | ||
| + | We resuspended and transformed the promoter+RBS plasmids from the distribution kit. They turn red when resuspended! Such a pretty color! | ||
| + | |||
| + | ===Tuesday=== | ||
| + | A lot of our reagents came in today! WOOOOOOOOo! | ||
| + | Since we’re having problems with transformation right now, we are troubleshooting our procedures to see if there’s anything wrong with our technique or protocols. We followed the iGEM standard transformation protocol and transformed our promoter+RBS plasmids again. Let’s hope they work! | ||
| + | |||
| + | [[File:ipredictfailure.png]] | ||
| + | |||
| + | ==Week 4 7/7 - 7/11== | ||
| + | ===Monday=== | ||
| + | Made overnight cultures of DH5a and IPTG+RBS to make competent cells/miniprep tomorrow. Inoculated MG1655 for gDNA and generic promoter + RBS for miniprep. Transformed plasmids containing mCherry+LVA (on cam) and GFP(on amp) into DH5a cells from distribution kit (mcherry: 2014 plate 3 well 12F and 2013 plate 3 well 11F, GFP 2014 plate 3 well 10k). Made 7ml overnight culture of MG1655. | ||
| + | |||
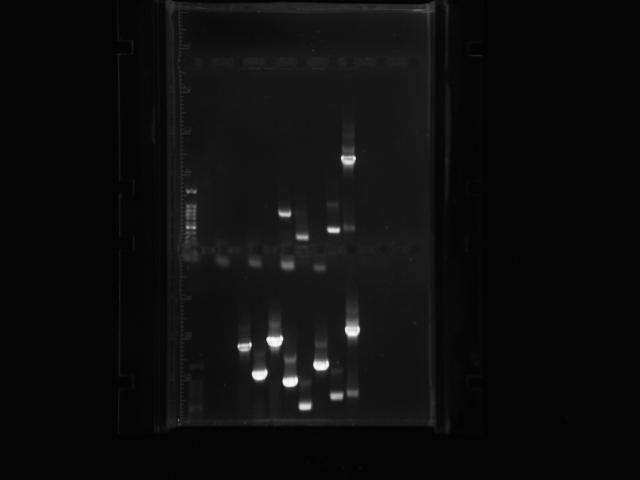
| + | ===Tuesday=== | ||
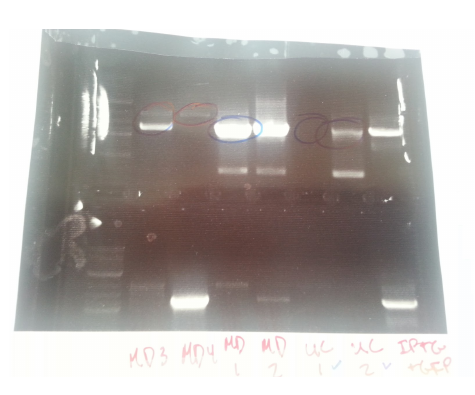
| + | Transformants all showed no growth (positive RFP control showed some growth but not red). Attempted to make DH5a competent cells. Ran mutator PCR products on a gel: | ||
| + | |||
| + | [[File:UChciago_wk4gel1.png]] | ||
| + | |||
| + | 1: Ladder | ||
| + | 2: Negative control | ||
| + | 3: mutD | ||
| + | 4: aroF | ||
| + | 5: MutH | ||
| + | 6: MutY | ||
| + | 7: MutL | ||
| + | 8: DinB | ||
| + | paroF successfully amplified at .3kb. | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Colonies are now growing on 7/7’s mCherry-LVA transformants plate and the positive control colonies have turned red. Transformed newly made competent cells with RFP plasmids to determine competency. PCR was performed with phusion and taq on the remaining mutators. | ||
| + | |||
| + | |||
| + | ===Thursday=== | ||
| + | Amplicons from yesterday were run on a gel: | ||
| + | |||
| + | [[File:UChciago_wk4gel2.png]] | ||
| + | |||
| + | P denotes phusion, T denotes Taq | ||
| + | #Ladder | ||
| + | #P1 -- 1.1kb | ||
| + | #P2 -- 0.7kb | ||
| + | #P3 -- N/A | ||
| + | #P4 -- N/A | ||
| + | #Ladder | ||
| + | #T1 -- 0.7kb | ||
| + | #T2 -- 0.7kb | ||
| + | #T3 -- 1.1kb | ||
| + | #T4 -- 0.7kb | ||
| + | #T5 -- 1.5.b | ||
| + | #Neg | ||
| + | |||
| + | no colonies for competency assay. Re-ran PCR on other mutators using taq and phusion. | ||
| + | |||
| + | ===Friday=== | ||
| + | 3 colonies on test plate (cells are not very competent). Miniprepped GFP plasmid ( cells borrowed from a different lab). | ||
| + | Ran gel from yesterday’s PCR: | ||
| + | |||
| + | [[File:UChciago_wk4gel3.png]] | ||
| + | |||
| + | Top gel: 1 ladder, 2 old primers mutD(taq), 3 new primers mutD(taq), 4 mutY old primers(taq), 5 mut Y new primers (phusion), 6 mutH old primers (taq), 7 mut H new (phusion), 8 mut L new (phusion), 9 negative control taq, 10 negative control phusion, 11 ladder | ||
| + | |||
| + | Bottom gel: 1 ladder, 2 mutL old (taq), 3 mutL new (taq) | ||
| + | |||
| + | ==Week 5 7/14 - 7/18== | ||
| + | |||
| + | ===Monday=== | ||
| + | We got the plasmids we ordered last week, including the 2 melA constructs (K193601 and K193602). We might not need them for a while, but always good to have them on hand. They came in agar stabs, so we restreaked them on plates. A bunch of our media was contaminated (again) so we had to remake all those. We also transformed our IPTG+GFP and our AroF+mCherry constructs, along with Cam, Kan, Tet, and Amp linearized backbones from the kit plate (other plasmids, cut in order to isolate the backbone). | ||
| + | |||
| + | [[File:ipredictfailure.png]] | ||
| + | |||
| + | (I’m calling this transformation.jpg now.) | ||
| + | |||
| + | ===Tuesday=== | ||
| + | Yep. Transformation of our ligated parts and the backbones seems to have failed. We left the plates in the incubator for one more night because these cells have an annoying habit of only growing after two days. At least our stab streaks were good: all 7 of the plasmids, including the important melA constructs, grew successfully, and were made into overnight cultures. | ||
| + | Oh hey! After a few hours more there is some growth on the backbone plates, except for the Tet backbone. Transformation efficiencies are pretty terrible, as is par for the course here. Seriously need to fix that. | ||
| + | We also redid our AroF+mCherry and our IPTG+GFP ligations with 3A assembly and the standard ligation procedure. Better work this time. We also miniprepped and fragment isolated more mCherry and GFP so that we have enough for the inevitable future redos. We’re also continuing work on determining the MIC for rifampicin for our fluctuation assays; this time growing them in liquid culture. | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Made glycerol stocks of most of Monday’s new plasmids, and miniprepped K193601 and K193602. Ran a bunch of PCRs to make sure we had our promoters. Didn’t get IPTG+GFP, but at least we got a little bit of AroF+mCherry. We also checked our MIC liquid cultures, and it looks like the MIC we got was about 8ug/mL. We also transformed our ligation products into our competent cells and competent cells from another lab. The other lab’s cells will most assuredly have better transformation efficiencies than our cells. | ||
| + | |||
| + | ===Thursday=== | ||
| + | Looks like our 3A fragments were cut incorrectly due to mislabeling somewhere along the line. Would probably explain why they failed this morning. | ||
| + | We can make our competent cells better, more competent. We have the technology. And buffer. Which we made today. | ||
| + | |||
| + | ===Friday=== | ||
| + | Plated some XL1-Blue cells to start making our competent cells. Hopefully these ones work better. Also tried ligating and transforming more mutators and promoters; hopefully we can do this without mislabeling things again. | ||
| + | (ipredictfailure.jpg) | ||
| + | |||
| + | ===Saturday=== | ||
| + | Made a culture of XL1-Blue cells with 250mL LB medium, growing cells to an optimal OD of 0.6 to transform. Looks like this will be going overnight, as growth is slow. Yesterday’s transformation products finally worked without incident: making them into overnight cultures so that we can have stocks in case of temporary incompetence. | ||
| + | |||
| + | ==Week 6 7/21 - 7/25== | ||
| + | ===Monday=== | ||
| + | We are spending the week attempting to clone our mutators into our IPTG-inducible promoter+GFP plasmid. We used up some of the fragment isolated mutators we had, so we gel purified more from wild type. The mutators and IPTG+GFP were appropriately digested (mutators with XbaI and PstI and IPTG+GFP with SpeI and PstI). We also performed a cell competency test on our cells (which are probably not competent, knowing our luck). | ||
| + | |||
| + | ===Tuesday=== | ||
| + | At the suggestion of our advisors, we are now dephosphorylating our plasmids before cloning to decrease background (which was a problem with our previous digestions). This should help a lot yay!! We dephosphorylated IPTG+GFP/S+P and gel purified the dephosphorylated digest. We cloned the mutators into IPTG+GFP using a ratio of 3:1 insert:vector in a 10uL reaction with 25ug of vector. We also submitted IPTG+GFP, mCherry+AroF, promoter+TyrR for sequencing. (We also made a ton of cam plates so no one freaks out the next time they’re transforming something and realizes there are no plates). This was a super late day--at lab past midnight=fun, fun, fun :) | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Our SOB is contaminated and an aliquot of SOC that was never used before contaminated. What is this madness?!?! More SOB was made. We also colony PCRed IPTG+GFP, mCherry+AroF, and promoter+TyrR in preparation for negative sequencing results (Always be prepared is one of our many mottos. We stole it from the Boy Scouts.). We also got Tyrosine!!!! | ||
| + | |||
| + | |||
| + | ===Thursday=== | ||
| + | Well, our sequencing results came back, and it appears we have MAJOR issues with mislabelling. While we do have IPTG+GFP confirmed, aroF and TyrR are not in anything | ||
| + | The plasmid mcherry is not even in anything. What was labeled as aroF+mCherry actually contains the general promoter, and one labeled promoter+TyrR is actually IPTG but without GFP!!! Are our mCherry and promoter plasmids mislabeled?! Or was it just the ligation products? The struggle is five real. We made overnight cultures of all the ligation products that looked promising based on the amount of colonies (our colony PCR did not work and this will allow us to keep moving forward while troubleshooting colony PCR): 4x IGMD (IPTG---GFP---MutD), 4x IGMH (IPTG---GFP---MutH). 4x IGMT (IPTG---GFP---MutT). 4x IGDam/IGD (IPTG---GFP---Dam). 4x IGUC (IPTG---GFP---Umu’C), 4x IGER (IPTG---GFP---EmrR). | ||
| + | |||
| + | ===Friday=== | ||
| + | Miniprepped all overnight cultures that were made yesterday of ligation constructs. We then PCRed them and chose constructs to submit for sequencing. | ||
| + | |||
| + | [[File:UChicago wk6prep1.png]] | ||
| + | |||
| + | [[File:UChicago Wk6prep2.png]] | ||
| + | |||
| + | [[File:UChicago wk6prep3.png]] | ||
| + | |||
| + | [[File:UChicago wk6prep4.png]] | ||
| + | |||
| + | ==Week 7 7/28 - 8/1== | ||
| + | ===Monday=== | ||
| + | This week we’re continuing lots of cloning. We were planning to make competent cells, but messed up the OD (which should be about 0.6) :(. Sadness. PCR’d out the AroF promoter and the TyrR gene and fragment isolated them. | ||
| + | |||
| + | [[File:UChicago wk7prep1.png]] | ||
| + | |||
| + | ===Tuesday=== | ||
| + | Re-inoculated new cells to retry making competent cells again. Hopefully they work this time. Did appropriate digestions on paroF, TyrR, the mCherry plasmid, and the IPTG-GFP mutator plasmid as we continue to clone all three different constructs (mutators, general promoter-TyrR, and paroF-mCherry). Did PCR’s using VF and VR2 primers to confirm the identity of the mCherry and constitutive promoter plasmids. However, mCherry looks possibly bad, so we ended up going ahead with cloning of only TyrR into the constitutive promoter plasmid. We got back sequencing results for three previous attempted ligations of Dam, EmrR, and MutT into IPTG-GFP, all of which look good. Yay, first real biobricks! | ||
| + | |||
| + | ===Wednesday=== | ||
| + | The cell line we’re using to make competent cells (XL1-Blu) that we’re trying to grow up is growing up faster than we expect, so we made a growth curve for how the OD changes over time. Did colony PCRs on attempted ligations of various mutators (Mut L, MutS, DinB, DnaE, MutY) into TyrR, but they all looked bad. Ligation fail. | ||
| + | |||
| + | [[File:UChicago wk7od.png]] | ||
| + | |||
| + | ===Thursday=== | ||
| + | We made competent cells today finally, and transformed some test plates to make sure they actually work. We redid the PCR to verify the mCherry plasmid, which again looked bad, so we’re just going to retransform mCherry just to be safe. | ||
| + | |||
| + | ===Friday=== | ||
| + | Success! Competent cells are actually competent after about 12 hours of growth. They look possibly pink, just as expected (since the plasmid they’re transformed with expresses RFP) | ||
| + | |||
| + | [[File:UChicago wk7culture1.png]] | ||
| + | |||
| + | [[File:UChicago wk7culture2.png]] | ||
| + | |||
| + | ==Week 8 8/4 - 8/8== | ||
| + | ===Monday=== | ||
| + | Resuspended and transformed mcherry+lva from the distribution kit again. We've been using this a lot and looks like we’re running pretty low on this! Let’s hope the transformation will succeed and our glycerol stock will be fine! Also transformed the sequence verified IPTG+GFP plasmid and those with EmrR, Dam and MutT for later use in the mutation rate analysis. | ||
| + | |||
| + | ===Tuesday=== | ||
| + | Transformation yesterday seems ok! Since there are a few colonies on the negative control, we restreaked the transformed colonies just to be sure. | ||
| + | And finally, the long-awaited MIC determination test was done today! As a preliminary experiment, we plated saturated, 1:100 and 1:10000 dilutions of XL1-Blue cells onto cam plates with various rif concentrations. | ||
| + | |||
| + | ===Wednesday=== | ||
| + | The restreaked colonies grew! Promptly checked the colonies by colony PCR and made overnight cultures of them for miniprepping. | ||
| + | |||
| + | Only less than 5 colonies grew on the 5ug/ml rif plates, and none on the other plates! This is unexpected, as there should be as many colonies as the plain LB plates in the lower rif concentration plates. Looks like there’s gonna be more rif experiments! | ||
| + | |||
| + | ===Thursday=== | ||
| + | As we accidentally added an extra RBS overhang in our AroF primer, we PCRed out AroF again using our new primer without the overhang. The gel looks good! Let's miniprep it! | ||
| + | |||
| + | And what??? 4 out of 7 overnight cultures from yesterday didn’t grow? And we have to pick the colonies again? We really need mcherry+lva… :( | ||
| + | But the IPTG+GFP plasmids look promising. Miniprepped the plasmids and ran them on a gel. Look at this beautiful gel! Now let’s cut it up for gel extraction! | ||
| + | |||
| + | [[File:UChicago wk8prep1.png]] | ||
| + | |||
| + | [[File:UChicago wk8prep2.png]] | ||
| + | |||
| + | ===Friday=== | ||
| + | What? Why didn’t the mcherry colonies grow again? :( Retransformed mcherry again. Ligated promoter and TyrR while waiting for transformation. | ||
| + | For the mutator plasmids, the remaining mutators were digested and ligated into the IPTG+GFP plasmids. Transformation results will be ready tomorrow! The ligated MutT and EmrR plasmids from earlier this week were miniprepped and stored. | ||
| + | |||
| + | ==Week 9 8/11 - 8/15== | ||
| + | ===Monday=== | ||
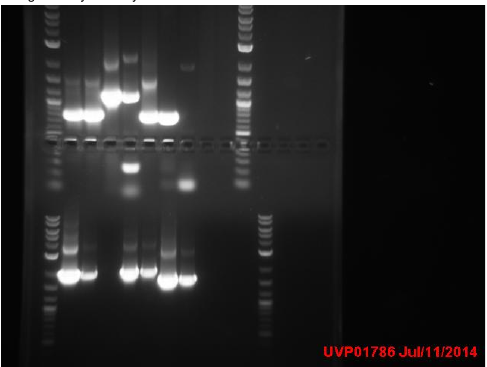
| + | It looks like one of the tubes labelled as “promoter plasmid” was actually IPTG+GFP. Whoopsy-daisy. Also transformed general promoter from kit plate. | ||
| + | A lot of the mutator transformation plates worked. Woot! We can now use them for cloning and stuff. Did colony PCR for a bunch of them and then ran on gels. MutH, MutY, and UmuC all gave good results. | ||
| + | |||
| + | ===Tuesday=== | ||
| + | Made glycerol stocks of mCherry. Miniprepped mCherry+Iva, then restriction digested and gel purified it. | ||
| + | Ran colony PCR reactions for MutS, MutL, and DinB. Ran gels for DnaE, UmuC (redo), and ParC (redo). The redos were done because redundancy is useful in science. Redundancy is redundant. Redundancy is also redundant. | ||
| + | Made a bunch of variable Rifampicin plates and diluted MG1655 into 200ul aliquots for mic determination. | ||
| + | |||
| + | Somewhere, in a deep, dark corner of the gel room, someone started making new TAE Buffer...but their mind was in physics mode instead of bio mode…*foreshadowing intensifies* | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Did colony PCR on a boatload (okay, it would have to be a really small boat) of IPTG+GFP so that we’d have plenty for future gels. Ran gels for MutS, MutL, and DinB. MutS and DinB gave some good results, but the two MutL gels didn’t work (one showed nothing, the other disintegrated). And so the buffer problems began. | ||
| + | Miniprepped MutH, ParC, UmuC, DnaE, and MutY. Twice. The first time failed miserably, since it turns out that one of the buffers (P2) was improperly made and had been giving us screwy results for a while. Some members wanted to keep it for some reason, so we just gave it a choke collar label that said “BAD” on it. The second miniprep attempt was successful with a better-behaved buffer. | ||
| + | Made glycerol stocks of promoter. Restriction digested and gel purified promoter. Ligated mCherry+paroF and promoter+TyrR and transformed products. | ||
| + | Plated independent cultures of MG1655 onto restrictive media and made dilutions (only up to around 10^6, though, nothing like that 1/10^30 homeopathy BS) | ||
| + | |||
| + | ===Thursday=== | ||
| + | Sent in a variety of mutators (MH, MY, UC, DE, and PC) for sequencing. Miniprepped DinB and MutS samples. Digested and ligated MutL and MutD with increased insert:vector ratio (now 5:1). Did Colony PCR on IGER3 and IG3 samples then ran on gels (used separate TAE buffer from the main stock, but the gels still failed anyways). | ||
| + | <strike>Ran Colony PCR and gels for mCherry+paroF and Promoter+TyrR</strike> Nevermind! The buffer is DEFINITELY bad, so there go the results for that! Time to do it all over again *eye twitches*. | ||
| + | Made more LB, rif, and dilution plates since the results from the previous ones were inconclusive. | ||
| + | |||
| + | ===Friday=== | ||
| + | Re-did the gels for mCherry+paroF and Promoter+TyrR. | ||
| + | Sent in DinB and MutS for sequencing. Transformed MutL, MutD, and IPTG+GFP into competent cells. It turns out that we didn’t need the IGER3 and IG3 overnight cultures that were made on Thursday, so they were thrown out. | ||
| + | Made new LB agar plates. Selective growth plates from the other day showed no growth, but the nonselective test plates showed reasonable amounts of growth. | ||
| + | |||
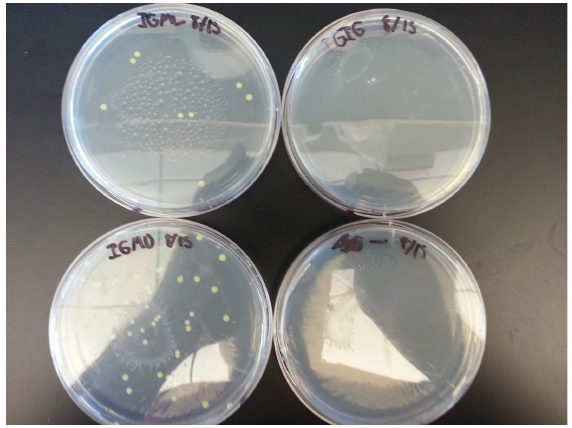
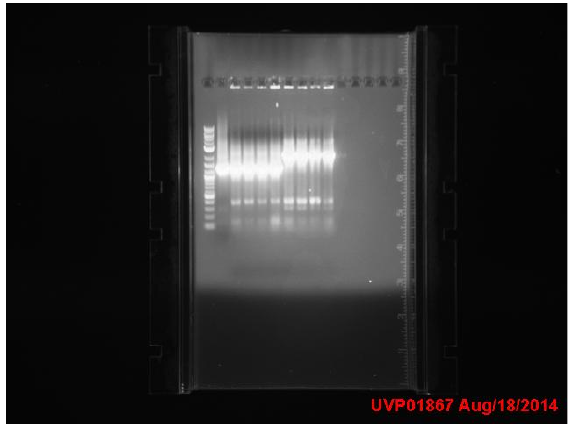
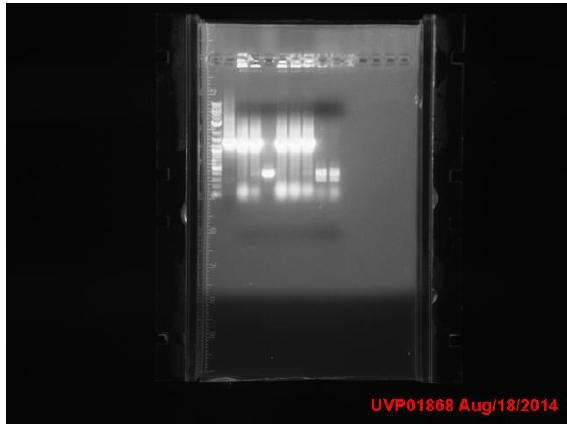
| + | ==Week 10 8/18 - 8/22== | ||
| + | Transformation results from 8/15 | ||
| + | for IPTG+GFP+mutL (IGML), IPTG+GFP+mutD (IGMD), IPTG+GFP (IGIG), and negative control: | ||
| + | |||
| + | [[File:UChicago wk10gel1.png]] | ||
| + | |||
| + | Low transformation efficiency suggests that the transformation was unsuccessful but mutD appears to be successful from the following colony PCR gel: | ||
| + | |||
| + | gel key: | ||
| + | Well 1: 4 µl of ladder | ||
| + | Well 2: 8 µl of IPTG+GFP | ||
| + | Well 3: Sample 1 | ||
| + | Well 4: Sample 2 | ||
| + | Well 5: Sample 3 | ||
| + | Well 6: Sample 4 | ||
| + | Well 7: Sample 5 | ||
| + | Well 8: Sample 6 | ||
| + | Well 9: Sample 7 | ||
| + | Well 10: Sample 8 | ||
| + | |||
| + | mutD gel: | ||
| + | |||
| + | [[File:UChicago wk10gel2.png]] | ||
| + | |||
| + | mutL gel: | ||
| + | |||
| + | [[File:UChicago wk10gel3.png]] | ||
| + | |||
| + | Transformation of IPTG+GFP+mutL appears to be unsuccessful but samples 4-8 of mutD appear to have the mutD insert as they have higher base pair count. | ||
| + | |||
| + | Quikchange of mutY and DinB was attempted but likely unsuccessful due to lack of any appropriate bands on the below gel and insufficient signal from sequencing | ||
| + | |||
| + | [[File:UChicago wk10gel4.png]] | ||
| + | |||
| + | 1: Ladder | ||
| + | 2: Neg control | ||
| + | 3: MutY | ||
| + | 4: DinB | ||
| + | Quikchange results transformed into XL1Blue cells. Overnight ligation of 10:1 TyrR:promoter(plasmid) attempted. Transformed ParC3, MutY7, DinB1, MutS8, MutH2, MutD5, and MutD8 into XL1Blue | ||
| + | Mutation rate assay: | ||
| + | Measured OD600 of XL1B wt overnight culture at 12:20: 1.617 (roughly 1.29x10^9 cells/mL) and made an inoculum with 5000 cells/mL in LB (with 1mM IPTG). Made 20 200ul aliquots of inoculum +IPTG and put into incubator at 1:16 pm. | ||
| + | |||
| + | ===Tuesday=== | ||
| + | Transformed mCherry+paroF, promoter+TyrR, mCherry+ligase, promoter+ligase, mCherry cut, promoter cut. No growth on mutY and DinB quikchange transformant plates. Attempted 2 more protocols for quikchange for mutY (2-step and 60oC annealing temperature). Both appeared to fail. | ||
| + | |||
| + | [[File:UChicago wk10gel5.png]] | ||
| + | |||
| + | 1: ladder | ||
| + | 2: 60C negative control | ||
| + | 3: 60C MutY product | ||
| + | 4: 2-Step negative control | ||
| + | 5: 2-Step MutY product | ||
| + | |||
| + | 2 mutD-positive colonies from 8/18 miniprepped. mutation rate assay: | ||
| + | 200ul cultures taken out of 37 degree shaker at 2:30 (~25 hrs), plated (16x) 200 ul on 80ug/mL rif plates as well as 200ul of 10^6 and 10^7 dilutions from 4 different overnight cultures on LB plates. The overnight culture of melA did not grow (the melA plasimid: BBa_K193602 has amp resistance rather than cam as listed on the registry). Inoculated overnight cultures at 5:00 in 5mL LB of XL1B, EmrR3, EmrR4, Dam). Resuspended 2 likely rif resistant colonies and plated on 80ug/mL rif plates to check if rif-resistant bacteria could grow at such a high concentration of rif. | ||
| + | |||
| + | ===Wednesday=== | ||
| + | PCRed out and gel-purified TyrR, paroF. Yesterday’s cut plasmids w/o ligase transformants yielded colonies, suggesting that the digestion and dephosphorylaion failed. Re-digested TyrR, paroF, mCherry, promoter and dephosphorylated mCherry and Promoter containing plasmids. Made overnight cultures of yesterday’s mutator transformants (ParC3, MutY7, DinB1, MutS8, MutH2, MutD5, and MutD8) . Redigested Umu’C and mutL and ligated into IPTG+GFP plasmid. melA overnight culture does not look any darker to the naked eye; a melA overnight culture was made with 250ug/mL tyrosine and 1mM IPTG in LB. Mutation rate assay: Dam master inoculum made of ~5000 cfu/mL from yesterday’s overnight culture, appropriate concentration of cam and 1mM IPTG which was then aliquotted to 19 200ul cultures. In addition, a rif80 plate streaked with likely rif-resistant colony from 8/18 have near-lawn growth showing that 80ug/mL rif is not too high a concentration to completely hinder growth. No growth in EmrR3 culture -reinoculated. In addition, XL1Blue overnight cultures were inoculated. | ||
| + | |||
| + | colony counts from XL1B wt test plates (200ul plated of the stated dilution): | ||
| + | {|class="wikitable" | ||
| + | |+Colony Counts | ||
| + | |- | ||
| + | !culture||10^6 dil||10^7 dil | ||
| + | |- | ||
| + | |1||61||8 | ||
| + | |- | ||
| + | |2||70||3 | ||
| + | |- | ||
| + | |3||63||1 | ||
| + | |- | ||
| + | |4||55||3 | ||
| + | |} | ||
| + | |||
| + | This suggests a cfu/mL concentration of ~5X10^7 was plated on selective plates (a low amount). | ||
| + | |||
| + | ===Thursday=== | ||
| + | Overnight cultures of ParC3, MutS, MutH2, DnaE, DinB1, MutY7, MutD5, and MutD8 miniprepped. Made additional overnight cultures of the above for glycerol stocks. mutY quikchange plates from 8/19 show a few colonies on the 2-step mutagenesis samples; these colonies have been used to inoculate overnight cultures. Ran 2-step quikchange protocol on DnaE, ParC, and mutH containing plasmids. Transformed yesterday’s IPTG+GFP+Umu’C/mutL ligation products as well as just IPTG+GFP into XL1Blue cells. paroF and TyrR were PCRed from genomic DNA and fragment isolated. paroF was digested and ligated into the mCherry plasmid, and TyrR into the promoter plasmid and the ligation products transfomed into XL1Blue. Mutation rate assay: 1 rif-resistant colony observed across 16 XL1Blue selective plates (plated on 8/19): likely a function of low plated cfu concentration. Significant growth on 2 rif80 plates platedwith rif-resistant bacteria from 8/19 further confirming that rif concentration is not an issue. Plated yesterday’s Dam 200ul culture aliquots on selective media as well as non-selective test plates (10^6,10^7,10^8 dilutions for three cultures). Made 20 mL culture aliquots of XL1Blue wt and put in 37 degree shaker (up from 200ul). Re-inoculated melA overnights for miniprep tomorrow (melA+tyrosine is not dark/barely grew and we want to sequence). | ||
| + | |||
| + | ===Friday=== | ||
| + | Made glycerol stocks of PC3, MS, MH2, DE, DB1, MY7, MD5, and MD8. Quikchange transformant plates from 8/21 show a few small colonies only on quikchanged mutH. Made colony suspensions of yesterday’s Umu’C and mutL ligation product transformants and ran on a gel after colony PCR: | ||
| + | Order: | ||
| + | Well 1: Ladder | ||
| + | Well 2: IPTG+GFP plasmid | ||
| + | Well 3: Sample 1 | ||
| + | Well 4: Sample 2 | ||
| + | Well 5: Sample 3 | ||
| + | Well 6: Sample 4 | ||
| + | Well 7: Sample 5 | ||
| + | Well 8: Sample 6 | ||
| + | Well 9: Sample 7 | ||
| + | Well 10: Sample 8 | ||
| + | MutL gel: | ||
| + | |||
| + | [[File:UChicago wk10gel6.png]] | ||
| + | |||
| + | unsuccessful. | ||
| + | Umu’C gel: | ||
| + | |||
| + | [[File:UChicago wk10gel7.png]] | ||
| + | |||
| + | No amplicons at the correct band (greater than 1kb). | ||
| + | |||
| + | Miniprepped mutY-quikchanged overnight cultures for sequencing. Quikchange transformants from yesterday still show only colonies(5) on the mutH plate. Transformed promoter+TyrR and aroF + mCherry ligation products yielded no colonies so re-ligated and retransformed. | ||
| + | |||
| + | |||
| + | ===Sunday=== | ||
| + | Mutation rate assay: no colonies on rif80 plates for Dam (16). Test plates showed inconsistent results (no colonies for culture 1 at any dilution, 1,0,4 colonies for culture 2 at 10^6 10^7, 10^8 dilutions, and 0, 2, 3 colonies for culture 3 at 10^6,10^7,10^8 dilutions). Plated XL1Blue 200ulx18 of the 2ml cultures on rif80 plates as well as 200ul of a 10^6 dilution on agar plates for 3 cultures. | ||
| + | |||
| + | Miniprepped melA but failed to achieve yield over 30ng/ul even with saturated 2x5ml cultures | ||
| + | |||
| + | ==Week 11 8/25 - 8/29== | ||
| + | ===Monday=== | ||
| + | We finally got our AroF+mCherry construct ligated in! Ran a colony PCR to verify these, and looks good so far. Made overnight cultures for miniprep tomorrow. | ||
| + | We also Dpn1-digested last week’s ParC and DnaE quickchange products and transformed them in. | ||
| + | |||
| + | [[File:ipredictfailure.png]] | ||
| + | |||
| + | Par for the course with this, really. We also ran a quickchange with ParC and DnaE again, this time under two different conditions: twice the amount of initial DNA and twice the amount of extension time (which is already too long). We transformed in the 2X DNA samples but the 2X time samples were not transformed until tomorrow. Also ran a colony PCR on last week’s MutL and UmuC ligation attempts: looks like half our UmuC colonies were successful, but all of MutL failed. | ||
| + | |||
| + | ===Tuesday=== | ||
| + | Miniprepped the mCherry+AroF constructs while admiring the culture’s pinkish color. Sent that and a UmuC ligation attempt in for sequencing. We also ran another PCR of the TPC and TDE quickchange products (the ones with a doubled extension time), and while one of the TDE samples had promise, none of the TPC samples showed anything. Still went ahead with all the transformations anyway, because these things tend to be wonky at times. We also miniprepped mMutH for sequencing and MelA for verification. | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Oh hey, a good sign! Monday’s mDnaE and mParC plates show a possible colony. Made overnight cultures of those and redid our MutS and MutY mutagenesis, with 2X DNA concentrations, since that seemed to have worked well. We transformed those in after Dpn1 digestion. Also sent in mMutH samples for sequencing and religated MutL, MutD, and DinB. We also ligated our promoter+TyrR construct and began a mutation rate assay of wild-type XL1-B cells. | ||
| + | |||
| + | ===Thursday=== | ||
| + | Transformed in the constructs we ligated yesterday. Also miniprepped yesterday’s overnight cultures, but not much growth was seen. Miniprep yields were disappointingly low, so there go those samples (i cry evry tym). At least we had colonies grow on our rifampicin plates. Made Dam overnight cultures and new IPTG stock. | ||
| + | Friday | ||
| + | Really good results for our TyrR construct! Looks like we’re actually starting to make progress! Colony PCR doesn’t seem to be working for this, but hopefully sequencing on Monday will tell us if we got it for certain. No growth on yesterday’s ligation plates, however, but we’re giving the plates one extra day to grow. We seem to be having the same problems that we had a few weeks before. Maybe it’s just a fluke. Also miniprepped yesterday’s overnight cultures. Yields were somewhat low as usual, but looks like we at least have enough for sequencing. Redoing MutS, ParC mutagenesis with a 68C extension temperature. | ||
| + | |||
| + | ===Saturday=== | ||
| + | Better results than usual! Apparently we had a colony on mMutY. We still need to get it sequenced, but maybe this one could work? Keeping our fingers crossed. We also miniprepped a bunch of mMutY samples. | ||
| + | |||
| + | ==Week 12 9/1 - 9/5== | ||
| + | ===Monday--Labor Day=== | ||
| + | Rikki was the only one who came in on a Federal Holiday. She ran colony PCR on Promoter+TyrR construct (but expected it not to work since for some reason it never amplifies the promoter well--she was right, you couldn’t see a band). She also made overnight cultures of the promoter+TyrR but only made two because she could not find more culture tubes. | ||
| + | |||
| + | ===Tueday=== | ||
| + | Miniprepped the two overnight cultures of Promoter+TyrR. We ran out of Promega buffers so we tried invitrogen, but the yields were too poor on one so only one sample was submitted. Guess this was a good thing because--spoiler--the results were negative. We also tried ethanol precipitation for miniprep yields which were too low to submit for sequencing and discovered experimentally that sometimes it works and sometimes it doesn’t. In addition to submitting Promoter+TyrR for sequencing, we also submitted mutagenized MutY and DnaE. DinB, MutD, MutL, UmuC, and IPTG+GFP were digested and the mutators were ligated to IPTG+GFP, this time as an overnight ligation at 4C (Promaga t4 DNA ligase calls for overnights to be done at 4C rather than 16C). We tried site-directed mutagenisis on ParC, MutS, MutY, and DnaE using turbo. Overnight cultures were made of the melanin plasmid, as well as IGDM and IGER (for rif assay). | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Transformed ligation products of IPTG+GFP+ DinB, MutD, MutL, UmuC. Assuming failure for Quickchange, trying again but using Turbo, Turbo hot start, Phusion hot start, Q5, and Q5 hot start polymerases side-by-side. Hopefully one will work this time :/ Promoter+TyrR sequencing came back negative today, so 12 overnight cultures were made (since colony PCR doesn’t work well for this construct). Rikki found a random IPTG+GFP+MutD plate in the cold room that looked promising (a lot of colonies, some visibly green, some white). Colony PCR was run on four green colonies and four white colonies. | ||
| + | |||
| + | ===Thursday=== | ||
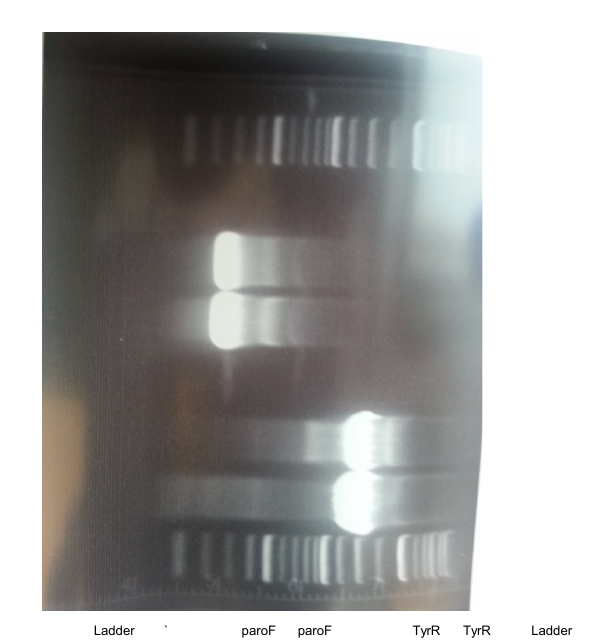
| + | Miniprepped Promoter+TyrR overnights. Rikki meshed together a new protocol using invitrogen buffers but following a lot of Promega’s timing and the yield’s are great--like 500ng/uL). The promoter+TyrR was digested with XbaI and PstI and run on a gel alongside digested promoter: | ||
| + | |||
| + | [[File:UChicago wk12gel1.png]] | ||
| + | |||
| + | ladder, promoter, samples 1-5, sample 12 | ||
| + | |||
| + | ladder, promoter, samples 6-11 | ||
| + | |||
| + | It looks like some of the samples have TyrR!!! (1541 bp) Sample 10 probably didn’t cut right. | ||
| + | Submitting samples 1, 3, 12, 7 for sequencing tomorrow morning. Looks promising!! | ||
| + | |||
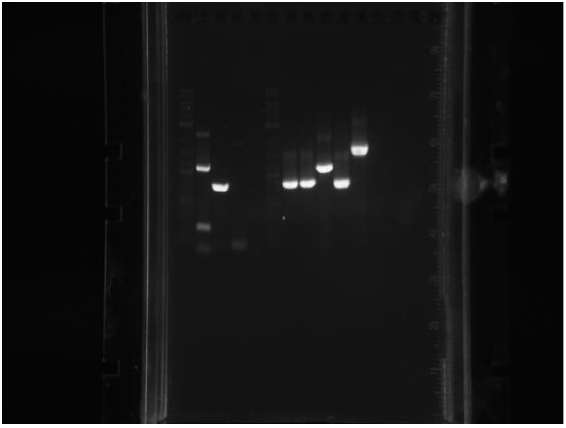
| + | Colony PCRed MutD: | ||
| + | |||
| + | [[File:UChicago wk12gel2.png]] | ||
| + | |||
| + | IPTG+GFP was in the first lane, but must not have been added correctly.We compared the bp size to our IPTG+GFP plasmid and it lookes like 5-8 are just IPTG+GFP (they were picked from green colonies). Samples 1-4 (picked white colonies) appear to have MutD, so they will be miniprepped and sent in for sequencing. | ||
| + | Plated EmrR overnight cultures on rif plates and dilutions on control plates (following the rif protocol) and made overnights of EmrR and Dam for the next batch of rif assays. | ||
| + | Miniprepped more IPTG+GFP for future ligations. | ||
| + | |||
| + | ===Friday=== | ||
| + | Submitted promising Promoter+TyrR and IPTG+GFP+MutD for sequencing. These results should be good!!! Three of the four mutators that were Quikchanged have colonies on the plates! It actually looks like Turbo doesn’t work as well as Q5 of Phusion, which is probably why our results for Quikchange have been pretty bad so far. Overnight cultures were made of these colonies. The ligations of IPTG+GFP+ DinB, MutD, MutL, UmuC don’t appear to have worked, and we’re probably going to scrap anything we don’t get soon. | ||
| + | |||
| + | ==Week 13 9/8 - 9/12== | ||
Summary | Summary | ||
| - | + | This was the week that several people (Rikki and Ryan B) were leaving lab and all our stuff suddenly started working last week, so it was an incredibly hectic week where we all stayed in lab until 2 or 3 in the morning every day. Summary of mutator progress by the end of this week (note to Derek/others: we should put this table up each week when available): | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | {|class="wikitable" |
| + | |+Mutator Summary | ||
| + | |- | ||
| + | !Mutator||Colony PCR?||Seq Verified?||Quikchange?||Overall progress | ||
| + | |- | ||
| + | |Dam||✓||✓||Not needed||Complete | ||
| + | |- | ||
| + | |DinB||X||X||F13V||N/A (may be able to get MIT else retry) | ||
| + | |- | ||
| + | |DnaE||✓||✓||E612K||Need sequencing primers to verify mutations! Ordered 9/9 | ||
| + | |- | ||
| + | |EmrR||✓||✓||Not needed||Complete | ||
| + | |- | ||
| + | |MutD||✓||✓||L73W, A164V||1st mutation in progress | ||
| + | |- | ||
| + | |MutH||✓||✓||E56A||Complete | ||
| + | |- | ||
| + | |MutL||X||X||E32K||N/A | ||
| + | |- | ||
| + | |MutS||✓||✓||G619D||Complete | ||
| + | |- | ||
| + | |MutT||✓||✓||Can’t use :(||N/A | ||
| + | |- | ||
| + | |MutY||✓||✓||V45A||Need sequencing primers to verify (start from bp 1)! ordered 9/10 | ||
| + | |- | ||
| + | |ParC||✓||✓||Y120H||Probably not doing due to the internal cut site. Eliot-Ryan can give it a shot though. | ||
| + | |- | ||
| + | |UmuC||X (MIT is sending it out. will deal with getting primers for it once we get it 9/11) it requires UmuD’ which has the first 24 amino acids of UmuD removed)||X||Y11A|| | ||
| + | |} | ||
| - | + | ===Monday=== | |
| - | + | We started PCRs to PCR out the mutators that have been successfully ligated (and Quikchanged when required) from the IPTG-GFP plasmid into the final ParoF-mCherry containing backbone we will actually carry out FREP in. For short, we call this ParoF-mCherry backbone ARM. We also continued various things, such as cloning DinB and MutL into IPTG-GFP, Quikchanging TyrR to make a mutated version of TyrR that has increased sensitivity and range of response to tyrosine, various already ligated things, and inoculating completely mutated Dam, EmrR, alongside wild-type XL1 Blu for the mutation rate assay. To test the promoter response to tyrosine repression, we also inoculated cells transformed with the AroF-TyrR construct into various concentrations of tyrosine (ranging from 0 to around 250 mg/L) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Tuesday=== | |
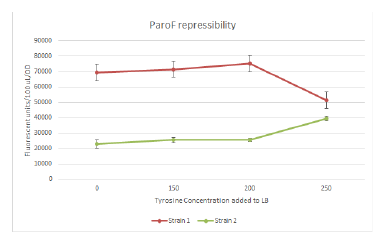
| - | + | Designed and ordered more oligos that we needed. We continued the Quikchanges and ligations from yesterday. Sequencing results came back, which allowed us to confirm MutS mutagenesis working. However, we’ll probably have to jump ship on ParC unfortunately, because there was an internal cut site that we only just now discovered/bothered to check, which means we only ligated in half of ParC :/ Sadness. We continued trying to PCR out, fragment isolate, and ligate the mutators (Dam EmrR, and MutH) into the ARM construct that we can use for FREP. Measured various OD’s from yesterday’s inoculation for the mutation rate assay, plating a humongous number of plates also (literally hundreds of plates). We also got plate reader data for the assay for promoter response to tyrosine repression, though it looks kinda sketch for one of the strains (appearing to be induced rather than repressed). | |
| - | + | [[File:UChicago_ryanb.jpg|300px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Happy birthday Ryan! ^This was at 2 AM on Wednesday morning since that’s how long we were in lab for. Stayed in lab till it was already Ryan’s birthday the next day, what a way to celebrate it XD | |
| - | + | ||
| - | + | [[File:UChicago_week13graph1.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | When the last people finally left lab at 3AM wednesday morning, we were all incredibly happy as we walked out the door. :D | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Wednesday=== | |
| - | + | We needed to make more competent cells (which we inoculated last night), but sadly overgrew it, so we’re gonna reinoculate and try again tomorrow. We transformed ARM ligations from yesterday, PCR’d to get linearized amp backbone we’ll need for the final functional TyrR construct (right now it’s on a chloramp backbone, but we need it on amp since the mutator construct will already be on chloramp). We ran colony PCR on the MutL and DinB ligations, both of which luckily showed lots of colonies. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Thursday=== | |
| - | + | Sad farewells :( Rikki and Ryan B left last night, so we are down to two people manning the lab. Eliot will be back next week and Kevin will be leaving after Friday. We miniprepped MutS and mutated TyrR (the latter of which needs to be sent in for sequencing verification of the mutagenesis) from yesterday. We also collected data for the mutation rate assay: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | {|class="wikitable" | |
| + | |+Colony count of XLB and Dam | ||
| + | |- | ||
| + | ! ||E||XLB||Dam | ||
| + | |- | ||
| + | |1||0 colonies||5 colonies||0 colonies | ||
| + | |- | ||
| + | |2||Near-lawn to lawn growth. Bad plate?||4 colonies||0 colonies | ||
| + | |- | ||
| + | |3||0 colonies||7 colonies||0 colonies | ||
| + | |- | ||
| + | |4||0 colonies||0 colonies||0 colonies | ||
| + | |- | ||
| + | |5||0 colonies||4 colonies||0 colonies | ||
| + | |- | ||
| + | |6||0 colonies||6 colonies||0 colonies | ||
| + | |- | ||
| + | |7||0 colonies||10 colonies||0 colonies | ||
| + | |- | ||
| + | |8||0 colonies||1 colonies||0 colonies | ||
| + | |- | ||
| + | |9||N/A||8 colonies||0 colonies | ||
| + | |- | ||
| + | |10||N/A||2 colonies||0 colonies | ||
| + | |- | ||
| + | |11||N/A||11 colonies||1 colonies | ||
| + | |- | ||
| + | |12||N/A||10 colonies||0 colonies | ||
| + | |- | ||
| + | |13||N/A||0 colonies||0 colonies | ||
| + | |- | ||
| + | |14||N/A||N/A||Bad plate? 1 cluster of colonies | ||
| + | |- | ||
| + | |15||N/A||N/A||0 colonies | ||
| + | |- | ||
| + | |16||N/A||N/A||0 colonies | ||
| + | |} | ||
| - | + | Looks like most of the mutator expressing plates have few colonies compared to wild type for whatever reason. Did colony PCR on the ARM and IPTG-GFP ligations from previous days, and found a few suitable colonies to send in (MutL specifically), though we need to do more colony PCR on some of the ligations (DinB, EmrR and DnaE specifically). | |
| - | + | We also have been collecting lysate from cells transformed with the various mutators we’ve been ligating, and ran it on an SDS-PAGE gel to confirm that the IPTG-GFP plasmids are properly expressing the mutators. We have a pic of the gel on the next day. | |
| - | + | Ladder | |
| - | + | #XLIB/IPTG (9/9) (7) | |
| - | + | #XLIB+IPTG (9/9) (1) | |
| - | + | #XLIB+IPTG (9/9) (5) | |
| - | + | #XLIB+IPTG (9/9) (6) | |
| - | + | #XLIB+IPTG (9/4) | |
| - | + | #EmrR+IPTG (9/9) (8) | |
| + | #EmrR+IPTG (9/9) (10) | ||
| + | #EmrR+IPTG (9/4) (1) | ||
| + | #EmrR+IPTG (9/4) (2) | ||
| + | #EmrR+IPTG (9/4) (3) | ||
| + | #EmrR/IPTG (9/9) (3) | ||
| + | #EmrR/IPTG (9/9) (4) | ||
| + | #EmrR/IPTG (9/4) (1) | ||
| + | #EmrR/IPTG (9/4) (2) | ||
| + | #Dam+IPTG (9/9) (9) | ||
| + | #Dam/IPTG (9/9) (2) | ||
| + | #Dam/IPTG (9/9) (11) | ||
| - | + | ===Friday=== | |
| - | + | We somehow lost our old mutated DnaE samples, so we went through the process of re-Quikchanging wild-type DnaE (alongside our newly ligated MutL). We also plan to ligate our sequence-verified, mutagenized MutS into ARM, so we set up the digests for that today, will need to ligate tomorrow. We ran more Colony PCRs of the ones that didn’t show any positives from before (see Thursday), and finally got more positives for EmrR and DamR, but still not for DinB (likely we’ll have to . We finished processing/imaging the protein gel we ran yesterday. A link here [[File:UChicago_wk13gel1.tif]]. | |
| - | + | Note: the protein gel is inverted so lane labels as listed from yesterday are backwards. The two rows should be identical; they’re just done at different protein concentrations to see which one would image better. We got some plasmids from Walker lab at MIT also today, which we put on nonselective LB plates for recovery. We may need these plasmids for PolB and UmuD’C, but on second thoughts (as you’ll see later), we ended up not actually needing them. We checked Dam and EmrR plates from the mutation rate assay that were plated on Thursday for growth, but there were few/no colonies, so we’ll wait one more day and see what happens as time works its magic. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Saturday=== | |
| - | + | Ryan S came in for a bit today to do various household chores. Checked plates we restreaked yesterday from glycerol stock to repeat the mutation rate assay, restreaked Walker lab plasmids from yesterday on selective plates (cam for the PolB plasmid and amp for the UmuD’C plasmid). He checked Dam and EmrR plates from the mutation rate assay that were plated on Thursday once again for growth and recorded down counts. Transformed various Quikchanges and ligations from earlier in the week we had been stalling an transforming (we wanted to transform a whole bunch of things at once to make things more efficient). Finally, made overnight cultures of the colony PCRs from Friday that looked good for EmrR and DamR that had been ligated into the ARM construct. | |
| - | + | ||
| - | + | ==Week 14 9/15 - 9/19== | |
| + | Summary | ||
| + | |||
| + | Running out of time/last final stretch: Everything’s coming together, but we still have a ton of work to do. | ||
| + | |||
| + | ===Sunday=== | ||
| + | Ryan S counted colonies on the nonselective XL1B plates and checked all the plates from yesterday. None of the transformation have large colonies so they were put for an additional day. | ||
| + | Miniprepped the overnight cultures of Dam and EmrR that actually grew. We have one of each! UmuD’C and PolB from the selective plates were innoculated and cultured in selective media. | ||
| + | Made glycerol stocks of the 4 mutators we have, and performed mutation rate analysis. Made a mastermix in LB, rif and IPTG at the right cell dilution and aliquotted out for overnight cultures. They’re now in the incubator and ready for tomorrow’s plating! | ||
| + | |||
| + | ===Monday=== | ||
| + | Checked all the transformation plates but none of them seems to have growth :( | ||
| + | Good news is, the overnight cultures of the mutators for mutation rate analysis (EmrR, MutS, MutH, Dam) have growth, so they were plated on LB plates and Rif plates when the right OD is reached. | ||
| + | DinB and UmuD’C were digested and ligated onto IPTG+GFP. PolB and UmuD’C were miniprepped and glycerol stocks were made. The miniprep sample concentration was really high - they were around 500 ng/ul! | ||
| + | |||
| + | ===Tuesday=== | ||
| + | UmuD’C and linearized amp backbone. Digested the UmuD’C, DinB and Dam and ligated them into IPTG+GFP with a 3:1 vector-insert ratio. AREmrR was sent in for sequencing and overnight cultures for ARDam was made for miniprepping. | ||
| + | Transformation plates from yesterday and the day before looked bad. The nonselective plates showed a lot of growth while the selective plates don’t. Seems like we need to do ligations and quickchange again… | ||
| + | |||
| + | Eliot then worked frantically to get something done! He quikchanged MutD, DnaE, MutL and TyrR. Then, he re-ligated UmuD’C and DinB into IPTG+GFP. | ||
| + | Sequencing results: | ||
| + | We have our mutated samples of mMutL, mMutY and mMutS and AroF+mCherry+mMutH! There was an extraneous mutation in TyrR that we used for all the ligations though. Fortunately, one of the templates (PT3) has a silent mutation. We could continue using this and make new quikchange templates of TyrR. | ||
| + | |||
| + | ===Wednesday=== | ||
| + | Sent in ARMEmrR and ARMDam for sequencing! However, overnights of ARMDam appear to have no growth...Made overnights again for miniprep and glycerol stocks. WE FOUND A GLYCEROL STOCK FOR THE ARM BACKBONE!! Quickly plated them for miniprep. | ||
| + | |||
| + | ===Thursday=== | ||
| + | ARMDam overnights finally have growth! Glycerol stocks made and miniprepped. Checked the mutation rate assay plates but not all of the selective plates have growth. We’ll check them again tomorrow! Made overnight cultures of XL1-B and JW-1316-1 for competent cells and ARM backbone for miniprepping. Also digested and ligated mMutY and mMutS into ARM backbone with 5:1 ratio. We ran out of competent cells so we’ll transform the products once the competent cells are made! | ||
| + | |||
| + | ===Friday=== | ||
| + | Miniprepped ARM backbone from yesterday’s overnight cultures. XL1-B and JW-1316-1 were cultured and competent cells were made. What a long day in the cold room! The cells were tested for their transformation efficiency using the iGEM kit. | ||
| + | Quikchanged MutL, MutD, DnaE and TyrR. Ligations from yesterday and these quikchanged products were transformed into competent Xl1-B and plated. | ||
| + | |||
| + | ===Saturday=== | ||
| + | Transformation efficiency of our competent cells is approximately 2X10^6. Hmmm it’s a bit lower than what we want but we’ll just go with it. | ||
| + | More overnight cultures of the mutators on IPTG+GFP were made for the mutation rate analysis! | ||
| + | |||
| + | ==Week 15 9/22 - 9/26== | ||
| + | Summary | ||
| - | + | It’s the final countdown! And we still don’t (and probably won’t) have the final FREP stuff good to go! (*Panicking intensifies*). But don’t FRET over FREP! McGuire and Eliot will do their best to finish what they can and lament what they cannot. | |
| - | + | ||
| - | + | ||
| - | * | + | |
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | ===Sunday=== |
| + | Ryan continued the Mutation Rate Assay protocol, checking ODs and making according overnight cultures. He also autoclaved various items and transformed Q1-5, P1-5, ARMY, ARMS, and ARMS1 for Eliot. | ||
| - | + | ===Monday=== | |
| - | + | Ryan checked ODs and plated cultures for the Mutation Rate Assay. He also started the final Mutation Rate Assay attempt, using XL1B with IPTG+GFP, Dam, IGER4, mMutH1, and mMutH2. McGuire also autoclaved a small boatload of micropipette tips. | |
| - | + | All work on cloning/mutagenesis was stopped today. End results: | |
| - | + | ||
| - | + | {|class="wikitable" | |
| - | + | |+Mutator Summary | |
| - | + | |- | |
| - | + | !Mutator||Colony PCR?||Seq Verified?||Quikchange?||Overall progress | |
| - | + | |- | |
| - | + | |Dam||✓||✓||Not needed||Complete | |
| - | + | |- | |
| - | + | |<strike>DinB</strike>||<strike>X</strike>||<strike>X</strike>||<strike>F13V</strike>||<strike>N/A (may be able to get MIT else retry)</strike> | |
| + | |- | ||
| + | |<strike>DnaE</strike>||<strike>✓</strike>||<strike>✓</strike>||<strike>E612K</strike>||<strike>Canceled at mutagenesis</strike> | ||
| + | |- | ||
| + | |EmrR||✓||✓||Not needed||Complete | ||
| + | |- | ||
| + | |<strike>MutD</strike>||<strike>✓</strike>||<strike>✓</strike>||<strike>L73W, A164V</strike>||<strike>Canceled at mutagenesis</strike> | ||
| + | |- | ||
| + | |MutH||✓||✓||E56A||Complete | ||
| + | |- | ||
| + | |<strike>MutL</strike>||<strike>X</strike>||<strike>X</strike>||<strike>E32K</strike>||<strike>Canceled at mutagenesis</strike> | ||
| + | |- | ||
| + | |<strike>MutS</strike>||<strike>✓</strike>||<strike>✓</strike>||<strike>G619D</strike>||<strike>Canceled at ARM ligation</strike> | ||
| + | |- | ||
| + | |<strike>MutT</strike>||<strike>✓</strike>||<strike>✓</strike>||<strike>Can’t use :(</strike>||<strike>N/A</strike> | ||
| + | |- | ||
| + | |MutY||✓||✓||V45A||Complete | ||
| + | |- | ||
| + | |<strike>ParC</strike>||<strike>✓</strike>||<strike>✓</strike>||<strike>Y120H</strike>||<strike>Probably not doing due to the internal cut site. Eliot-Ryan can give it a shot though.</strike> | ||
| + | |- | ||
| + | |<strike>UmuC</strike>||<strike>X (MIT is sending it out. will deal with getting primers for it once we get it 9/11) it requires UmuD’ which has the first 24 amino acids of UmuD removed)</strike>||<strike>X</strike>||<strike>Y11A</strike>||<strike>Cancelled at IG ligation, time constraints</strike> | ||
| + | |} | ||
| - | + | Eliot started concentrating on the tyrosine assay. Since ligating TyrR into ARM would take too long and be subject to setbacks, he used only the ARM backbone with endogenous, wild-type TyrR in DH5a. Eliot transformed TyrR into DH5A cells today, growth in LB with TyrR to come on Tuesday. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Tuesday=== | |
| - | + | Ryan continued the Mutation Rate Assay protocol, checking ODs and making according overnight cultures. Also checked the nonselective plates from Monday. | |
| - | + | Eliot delayed transformation of the ARM backbone delayed since either he goofed and added DNA to the negative control or contamination or plates were bad or something. Ergo, Eliot decided to redo the transformation, making sure to grab a plate from a different batch this time. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Wednesday=== | |
| - | + | Ryan checked ODs and plated cultures for the Mutation Rate Assay and made 40 more rif plates. | |
| - | + | Eliot’s transformation went much better this time. Negative control looked weird: there might be some growth but the agar in this plate was weird, so it could have been the agar looking like that. ARM plate actually showed good growth, however, compared to the other plate. Eliot made the tyrosine cultures and added samples of 3 colonies to each. Needed to match the tyrosine concentrations, though. Eugene suggested starting from a controlled concentration and diluting it down, so trying that. Planning to pick 3 colonies, add them to different concentrations of tyrosine. | |
| - | + | Concentrations: 0-500mg/L, in 50mg/L intervals (50, 100, 150…) | |
| - | + | 3 colonies picked, suspended in 35uL H2O each. | |
| - | + | 2uL suspended colonies put in 5uL LB/Tyr | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Thursday=== | |
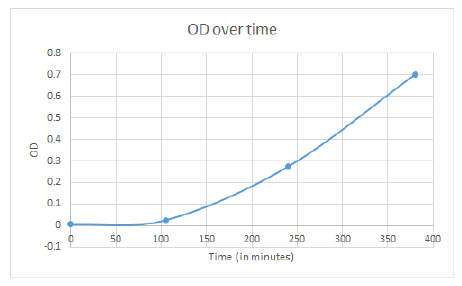
| - | + | Eliot took cultures at 12:05 PM, total incubation period is 16 hours. Successful growth occurred in most cultures, with the exception of 2 vials: should not affect results too much if data is consistent across all 3 colonies, as the vials correspond to different colonies and are not in the range of 200-300mg/L tyrosine, which appears to be the point at which AroF is repressed judging from the previous assay. He loaded approx. 200 µL culture into wells of 96-well plate. Rows A, B, and C corresponded to colonies 1, 2, and 3. Columns 1-12 were: 0=LB, no cells, 1, 0mg/uL tyrosine, 2=50mg/L tyr, 3= 100mg/L tyr, … 12= 500mg/L tyr. Measured 570-610mm. | |
| - | + | Ryan checked nonselective plates for the Mutation Rate Assay. All fluoresced (yay)! | |
| - | + | Friday | |
| - | + | Ryan and Eliot were both busy, so no one went into the lab on this day of Fries. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ===Saturday=== | |
| - | + | Ryan stared across the barren wasteland lab, wondering if it was all worth it. Had all the blood, sweat, and tears been shed for this? This...end? It seemed that this experiment shall go out with a whimper rather than a bang...Ah, well. | |
| - | + | Ryan checked the selective plates for the Mutation Rate Assay. All of these had fluorescing colonies as well. | |
| - | + | McGuire headed to the Museum of Science and Industry to drink some blue lemonade for free whilst standing inside of a manmade mini-tornado. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <html></div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 03:26, 18 October 2014
Lab Notebook
See Media:UChicago_LabNotebookSummary.pdf for the original document.
Week 1 6/16 - 6/20
Monday
First day of lab! We plated some DH5-alpha competent cells in preparation for gDNA extraction for our PCRs and future transformations. One person showed up (Kevin yayyyyy), then left.
Tuesday
We attempted to make LB plates. We had fun watching it boiling over since we left it in the hot plate too long and it spilled everywhere. We did clean-up time afterwards! In the future, we will remember to to heat for only 1 min to dissolve everything except agar. Agar will dissolve during autoclave because science is cool. So exciting! :D We then remade the LB for funsies :D We did a few other things to start things up like making SOB, CCMB80 competent cell buffer, and antibiotic plates.
Wednesday
MOAR BUFFERS! Qiagen buffers are expensive so we spent the day making bootleg Qiagen miniprep buffers (sorry Qiagen, you had your chance). Then, we contemplated life for the rest of the day. *take depressing pictures here*
- take pictures of buffers*
Thursday
Made competent cells. Stuck hands in liquid nitrogen. Burnt our hands off (no pics because you need hands to use a camera, duh). EVEN MOAR BUFFERS We also went to a biotech company exhibit and grabbed a lot of free samples! (and food of course) We also extracted gDNA from DH5a to PCR out our mutator genes. We also learned not to use the free 1.5mL centrifuge tubes from last year, and to not smell our microcentrifuge anymore. Stock strains MG1655 and delta-tyrR came in from the Yale E Coli Stock Center. Yay! So exciting! Kevin was so excited that he burnt his hair trying to plate them. XD
We made competent cells (MY CRYSTAL BALL SAYS: THEY WILL FAIL). Kevin BS’d the gDNA and got some somehow but also made the centrifuge stink like chloroform/phenol. Fun XD
This company (name concealed) gave us some cool centrifuge tubes last year that warp when we spin them down. XD So we’re buying new ones. $$$ They’re wrong- money DOES grows on trees! AND we have lots of trees around here! Even better! We ran overnight PCRs, one with more and one with less cycles.
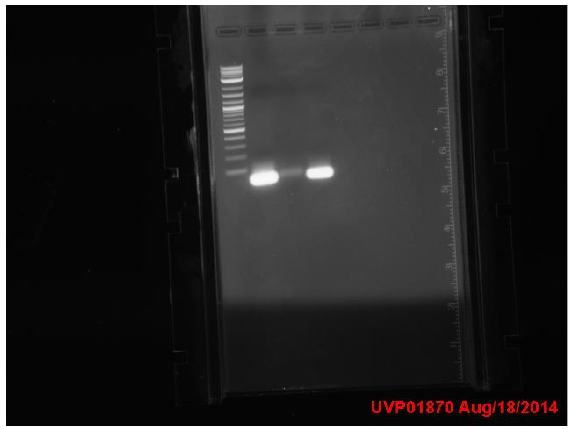
Friday
PCR-ed out 9/12 mutators. The lengths of almost all of them were verified to be correct!!! So much happy!!! Yes!!!! Yay!!!! The PCR with less cycles (bottom half of the gel) worked better so we’ll stick with that in the future. We also digested backbones that have the RBS we will use and ligate to all the mutators. What a good day!
Week 2 6/23 - 6/27
Monday
We tested our competent cells using the transformation efficiency kit. They didn’t work XD But that’s okay, we like making competent cells. It’s strangely exciting to pipette things lots of times :D
Also, we fragment isolated the mutations and then did the restriction digests on the PCR products. The concentrations of the genes are fairly alright, about 20-50 ng/microL. We ran a gel of AroF to verify PCR of the gene, but unfortunately we did not get a band XD. That’s okay, life goes on even if your gels fail!!! XD IT’S summer time! Rainbows!
We gel extracted the mutators that were digested last week. Fun. Got high contamination. But contamination is good because what doesn’t kill your experiments makes them stronger right ? Yes! Concentrations are listed below XD
| Sample | Conc (ng/microL) | 260/280 | 260/230 |
|---|---|---|---|
| ParC | 30.2 | 1.78 | 0.25 |
| DinB | 55.8 | 1.70 | 0.42 |
| MutS | 21.3 | 1.86 | 0.18 |
| Dam | 37.0 | 1.68 | 0.18 |
| MutT | 29.9 | 1.61 | 0.09 |
| Umu’C | 24.5 | 1.80 | 0.05 |
| EmrR | 36.8 | 1.71 | 0.08 |
| DnaE | 31.1 | 1.53 | 0.32 |
Tuesday
NOTHING WORKED :)))))!!!!!!!! We drank our worries away XD XD XD. We drunkenly made glycerol stocks of the two strains from Yale that came last Thursday.
- hic* guuysh guysh we should *burrrrp* totally ligate a rBs to a vecter *hi*c
Alcohol is poison. (Delicious, delicious poison. And we are Mithridates.)
Unfortunately, our cells weren’t competent when we tested them again :( We went over the protocol and made sure we’ll have everything down for next time XD. Or rather, fortunately! We get to make more! XD Soooo stoked! We fragment isolated and gel purified our promoter plasmids. For our project planning, we met with our faculty advisors Mike and Jennifer. The meeting was super, super, super productive and educational! We could consider inserting a polylinker into our plasmids to reduce a lot of steps XD Polylinkers are fun!
Wednesday
We purified the 8 mutators that we PCR-ed out. As for AroF and the three mutators that didn’t work, we tried PCR with Phusion, but unfortunately it didn’t work again. Yay! After digesting our RBS and ligating it with our previous cut promoter plasmids, we also did 3A assembly in parallel for RBS and promoter. Yay! More amp and tet plates were made! Yayyy!
- picture of plates”
Thursday
We had issues with SOC contamination today - We had to make a new batch for more transformations. We also tried transformation with the efficiency kit again, making sure we were following each step as it was written on our protocol. Friday Transformation of our 3A assembly colonies didn’t work, but the transformation of mcherry plasmids worked! This means that there’s nothing wrong with our transformation protocol. We will look into our digestion and ligation steps. We <3 troubleshooting! All part of the scientific process XD! Here’s a joke XD: What do you call it when McDonald sells cherries? mCherry! HAHAHA XD!
We made more chloramphenicol plates again! Yay!
Week 3 6/30 - 7/4
Monday
We resuspended and transformed the promoter+RBS plasmids from the distribution kit. They turn red when resuspended! Such a pretty color!
Tuesday
A lot of our reagents came in today! WOOOOOOOOo! Since we’re having problems with transformation right now, we are troubleshooting our procedures to see if there’s anything wrong with our technique or protocols. We followed the iGEM standard transformation protocol and transformed our promoter+RBS plasmids again. Let’s hope they work!
Week 4 7/7 - 7/11
Monday
Made overnight cultures of DH5a and IPTG+RBS to make competent cells/miniprep tomorrow. Inoculated MG1655 for gDNA and generic promoter + RBS for miniprep. Transformed plasmids containing mCherry+LVA (on cam) and GFP(on amp) into DH5a cells from distribution kit (mcherry: 2014 plate 3 well 12F and 2013 plate 3 well 11F, GFP 2014 plate 3 well 10k). Made 7ml overnight culture of MG1655.
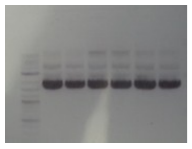
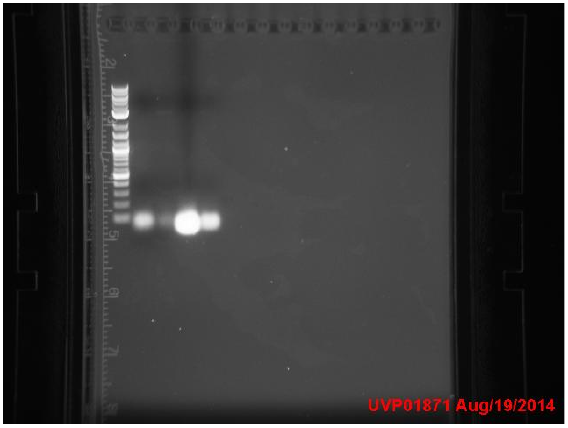
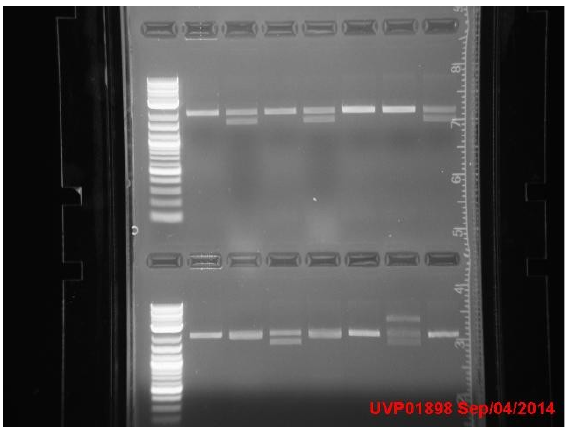
Tuesday
Transformants all showed no growth (positive RFP control showed some growth but not red). Attempted to make DH5a competent cells. Ran mutator PCR products on a gel:
1: Ladder 2: Negative control 3: mutD 4: aroF 5: MutH 6: MutY 7: MutL 8: DinB paroF successfully amplified at .3kb.
Wednesday
Colonies are now growing on 7/7’s mCherry-LVA transformants plate and the positive control colonies have turned red. Transformed newly made competent cells with RFP plasmids to determine competency. PCR was performed with phusion and taq on the remaining mutators.
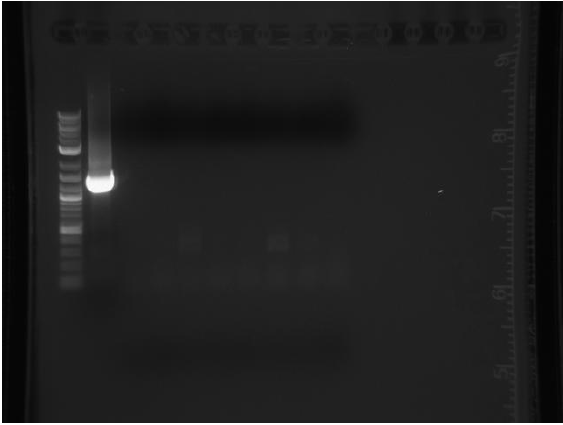
Thursday
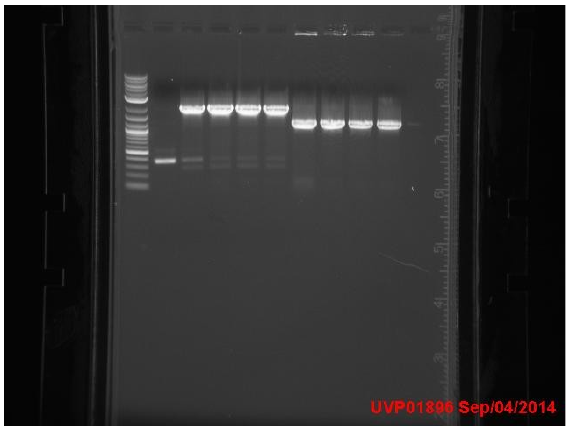
Amplicons from yesterday were run on a gel:
P denotes phusion, T denotes Taq
- Ladder
- P1 -- 1.1kb
- P2 -- 0.7kb
- P3 -- N/A
- P4 -- N/A
- Ladder
- T1 -- 0.7kb
- T2 -- 0.7kb
- T3 -- 1.1kb
- T4 -- 0.7kb
- T5 -- 1.5.b
- Neg
no colonies for competency assay. Re-ran PCR on other mutators using taq and phusion.
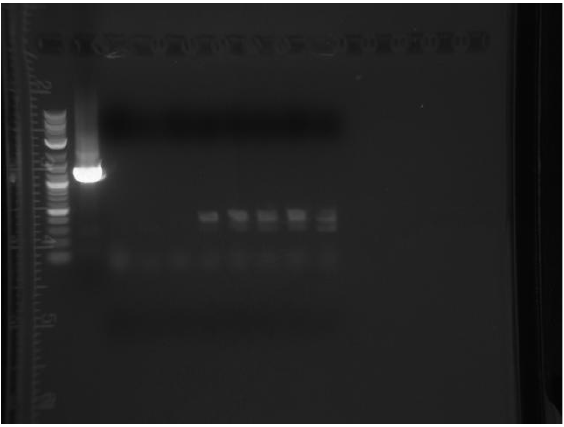
Friday
3 colonies on test plate (cells are not very competent). Miniprepped GFP plasmid ( cells borrowed from a different lab). Ran gel from yesterday’s PCR:
Top gel: 1 ladder, 2 old primers mutD(taq), 3 new primers mutD(taq), 4 mutY old primers(taq), 5 mut Y new primers (phusion), 6 mutH old primers (taq), 7 mut H new (phusion), 8 mut L new (phusion), 9 negative control taq, 10 negative control phusion, 11 ladder
Bottom gel: 1 ladder, 2 mutL old (taq), 3 mutL new (taq)
Week 5 7/14 - 7/18
Monday
We got the plasmids we ordered last week, including the 2 melA constructs (K193601 and K193602). We might not need them for a while, but always good to have them on hand. They came in agar stabs, so we restreaked them on plates. A bunch of our media was contaminated (again) so we had to remake all those. We also transformed our IPTG+GFP and our AroF+mCherry constructs, along with Cam, Kan, Tet, and Amp linearized backbones from the kit plate (other plasmids, cut in order to isolate the backbone).
(I’m calling this transformation.jpg now.)
Tuesday
Yep. Transformation of our ligated parts and the backbones seems to have failed. We left the plates in the incubator for one more night because these cells have an annoying habit of only growing after two days. At least our stab streaks were good: all 7 of the plasmids, including the important melA constructs, grew successfully, and were made into overnight cultures. Oh hey! After a few hours more there is some growth on the backbone plates, except for the Tet backbone. Transformation efficiencies are pretty terrible, as is par for the course here. Seriously need to fix that. We also redid our AroF+mCherry and our IPTG+GFP ligations with 3A assembly and the standard ligation procedure. Better work this time. We also miniprepped and fragment isolated more mCherry and GFP so that we have enough for the inevitable future redos. We’re also continuing work on determining the MIC for rifampicin for our fluctuation assays; this time growing them in liquid culture.
Wednesday
Made glycerol stocks of most of Monday’s new plasmids, and miniprepped K193601 and K193602. Ran a bunch of PCRs to make sure we had our promoters. Didn’t get IPTG+GFP, but at least we got a little bit of AroF+mCherry. We also checked our MIC liquid cultures, and it looks like the MIC we got was about 8ug/mL. We also transformed our ligation products into our competent cells and competent cells from another lab. The other lab’s cells will most assuredly have better transformation efficiencies than our cells.
Thursday
Looks like our 3A fragments were cut incorrectly due to mislabeling somewhere along the line. Would probably explain why they failed this morning. We can make our competent cells better, more competent. We have the technology. And buffer. Which we made today.
Friday
Plated some XL1-Blue cells to start making our competent cells. Hopefully these ones work better. Also tried ligating and transforming more mutators and promoters; hopefully we can do this without mislabeling things again. (ipredictfailure.jpg)
Saturday
Made a culture of XL1-Blue cells with 250mL LB medium, growing cells to an optimal OD of 0.6 to transform. Looks like this will be going overnight, as growth is slow. Yesterday’s transformation products finally worked without incident: making them into overnight cultures so that we can have stocks in case of temporary incompetence.
Week 6 7/21 - 7/25
Monday
We are spending the week attempting to clone our mutators into our IPTG-inducible promoter+GFP plasmid. We used up some of the fragment isolated mutators we had, so we gel purified more from wild type. The mutators and IPTG+GFP were appropriately digested (mutators with XbaI and PstI and IPTG+GFP with SpeI and PstI). We also performed a cell competency test on our cells (which are probably not competent, knowing our luck).
Tuesday
At the suggestion of our advisors, we are now dephosphorylating our plasmids before cloning to decrease background (which was a problem with our previous digestions). This should help a lot yay!! We dephosphorylated IPTG+GFP/S+P and gel purified the dephosphorylated digest. We cloned the mutators into IPTG+GFP using a ratio of 3:1 insert:vector in a 10uL reaction with 25ug of vector. We also submitted IPTG+GFP, mCherry+AroF, promoter+TyrR for sequencing. (We also made a ton of cam plates so no one freaks out the next time they’re transforming something and realizes there are no plates). This was a super late day--at lab past midnight=fun, fun, fun :)
Wednesday
Our SOB is contaminated and an aliquot of SOC that was never used before contaminated. What is this madness?!?! More SOB was made. We also colony PCRed IPTG+GFP, mCherry+AroF, and promoter+TyrR in preparation for negative sequencing results (Always be prepared is one of our many mottos. We stole it from the Boy Scouts.). We also got Tyrosine!!!!
Thursday
Well, our sequencing results came back, and it appears we have MAJOR issues with mislabelling. While we do have IPTG+GFP confirmed, aroF and TyrR are not in anything The plasmid mcherry is not even in anything. What was labeled as aroF+mCherry actually contains the general promoter, and one labeled promoter+TyrR is actually IPTG but without GFP!!! Are our mCherry and promoter plasmids mislabeled?! Or was it just the ligation products? The struggle is five real. We made overnight cultures of all the ligation products that looked promising based on the amount of colonies (our colony PCR did not work and this will allow us to keep moving forward while troubleshooting colony PCR): 4x IGMD (IPTG---GFP---MutD), 4x IGMH (IPTG---GFP---MutH). 4x IGMT (IPTG---GFP---MutT). 4x IGDam/IGD (IPTG---GFP---Dam). 4x IGUC (IPTG---GFP---Umu’C), 4x IGER (IPTG---GFP---EmrR).
Friday
Miniprepped all overnight cultures that were made yesterday of ligation constructs. We then PCRed them and chose constructs to submit for sequencing.
Week 7 7/28 - 8/1
Monday
This week we’re continuing lots of cloning. We were planning to make competent cells, but messed up the OD (which should be about 0.6) :(. Sadness. PCR’d out the AroF promoter and the TyrR gene and fragment isolated them.
Tuesday
Re-inoculated new cells to retry making competent cells again. Hopefully they work this time. Did appropriate digestions on paroF, TyrR, the mCherry plasmid, and the IPTG-GFP mutator plasmid as we continue to clone all three different constructs (mutators, general promoter-TyrR, and paroF-mCherry). Did PCR’s using VF and VR2 primers to confirm the identity of the mCherry and constitutive promoter plasmids. However, mCherry looks possibly bad, so we ended up going ahead with cloning of only TyrR into the constitutive promoter plasmid. We got back sequencing results for three previous attempted ligations of Dam, EmrR, and MutT into IPTG-GFP, all of which look good. Yay, first real biobricks!
Wednesday
The cell line we’re using to make competent cells (XL1-Blu) that we’re trying to grow up is growing up faster than we expect, so we made a growth curve for how the OD changes over time. Did colony PCRs on attempted ligations of various mutators (Mut L, MutS, DinB, DnaE, MutY) into TyrR, but they all looked bad. Ligation fail.
Thursday
We made competent cells today finally, and transformed some test plates to make sure they actually work. We redid the PCR to verify the mCherry plasmid, which again looked bad, so we’re just going to retransform mCherry just to be safe.
Friday
Success! Competent cells are actually competent after about 12 hours of growth. They look possibly pink, just as expected (since the plasmid they’re transformed with expresses RFP)
Week 8 8/4 - 8/8
Monday
Resuspended and transformed mcherry+lva from the distribution kit again. We've been using this a lot and looks like we’re running pretty low on this! Let’s hope the transformation will succeed and our glycerol stock will be fine! Also transformed the sequence verified IPTG+GFP plasmid and those with EmrR, Dam and MutT for later use in the mutation rate analysis.
Tuesday
Transformation yesterday seems ok! Since there are a few colonies on the negative control, we restreaked the transformed colonies just to be sure. And finally, the long-awaited MIC determination test was done today! As a preliminary experiment, we plated saturated, 1:100 and 1:10000 dilutions of XL1-Blue cells onto cam plates with various rif concentrations.
Wednesday
The restreaked colonies grew! Promptly checked the colonies by colony PCR and made overnight cultures of them for miniprepping.
Only less than 5 colonies grew on the 5ug/ml rif plates, and none on the other plates! This is unexpected, as there should be as many colonies as the plain LB plates in the lower rif concentration plates. Looks like there’s gonna be more rif experiments!
Thursday
As we accidentally added an extra RBS overhang in our AroF primer, we PCRed out AroF again using our new primer without the overhang. The gel looks good! Let's miniprep it!
And what??? 4 out of 7 overnight cultures from yesterday didn’t grow? And we have to pick the colonies again? We really need mcherry+lva… :( But the IPTG+GFP plasmids look promising. Miniprepped the plasmids and ran them on a gel. Look at this beautiful gel! Now let’s cut it up for gel extraction!
Friday
What? Why didn’t the mcherry colonies grow again? :( Retransformed mcherry again. Ligated promoter and TyrR while waiting for transformation. For the mutator plasmids, the remaining mutators were digested and ligated into the IPTG+GFP plasmids. Transformation results will be ready tomorrow! The ligated MutT and EmrR plasmids from earlier this week were miniprepped and stored.
Week 9 8/11 - 8/15
Monday
It looks like one of the tubes labelled as “promoter plasmid” was actually IPTG+GFP. Whoopsy-daisy. Also transformed general promoter from kit plate. A lot of the mutator transformation plates worked. Woot! We can now use them for cloning and stuff. Did colony PCR for a bunch of them and then ran on gels. MutH, MutY, and UmuC all gave good results.
Tuesday
Made glycerol stocks of mCherry. Miniprepped mCherry+Iva, then restriction digested and gel purified it. Ran colony PCR reactions for MutS, MutL, and DinB. Ran gels for DnaE, UmuC (redo), and ParC (redo). The redos were done because redundancy is useful in science. Redundancy is redundant. Redundancy is also redundant. Made a bunch of variable Rifampicin plates and diluted MG1655 into 200ul aliquots for mic determination.
Somewhere, in a deep, dark corner of the gel room, someone started making new TAE Buffer...but their mind was in physics mode instead of bio mode…*foreshadowing intensifies*
Wednesday
Did colony PCR on a boatload (okay, it would have to be a really small boat) of IPTG+GFP so that we’d have plenty for future gels. Ran gels for MutS, MutL, and DinB. MutS and DinB gave some good results, but the two MutL gels didn’t work (one showed nothing, the other disintegrated). And so the buffer problems began. Miniprepped MutH, ParC, UmuC, DnaE, and MutY. Twice. The first time failed miserably, since it turns out that one of the buffers (P2) was improperly made and had been giving us screwy results for a while. Some members wanted to keep it for some reason, so we just gave it a choke collar label that said “BAD” on it. The second miniprep attempt was successful with a better-behaved buffer. Made glycerol stocks of promoter. Restriction digested and gel purified promoter. Ligated mCherry+paroF and promoter+TyrR and transformed products. Plated independent cultures of MG1655 onto restrictive media and made dilutions (only up to around 10^6, though, nothing like that 1/10^30 homeopathy BS)
Thursday
Sent in a variety of mutators (MH, MY, UC, DE, and PC) for sequencing. Miniprepped DinB and MutS samples. Digested and ligated MutL and MutD with increased insert:vector ratio (now 5:1). Did Colony PCR on IGER3 and IG3 samples then ran on gels (used separate TAE buffer from the main stock, but the gels still failed anyways).
Ran Colony PCR and gels for mCherry+paroF and Promoter+TyrR Nevermind! The buffer is DEFINITELY bad, so there go the results for that! Time to do it all over again *eye twitches*.
Made more LB, rif, and dilution plates since the results from the previous ones were inconclusive.
Friday
Re-did the gels for mCherry+paroF and Promoter+TyrR. Sent in DinB and MutS for sequencing. Transformed MutL, MutD, and IPTG+GFP into competent cells. It turns out that we didn’t need the IGER3 and IG3 overnight cultures that were made on Thursday, so they were thrown out. Made new LB agar plates. Selective growth plates from the other day showed no growth, but the nonselective test plates showed reasonable amounts of growth.
Week 10 8/18 - 8/22
Transformation results from 8/15 for IPTG+GFP+mutL (IGML), IPTG+GFP+mutD (IGMD), IPTG+GFP (IGIG), and negative control:
Low transformation efficiency suggests that the transformation was unsuccessful but mutD appears to be successful from the following colony PCR gel:
gel key: Well 1: 4 µl of ladder Well 2: 8 µl of IPTG+GFP Well 3: Sample 1 Well 4: Sample 2 Well 5: Sample 3 Well 6: Sample 4 Well 7: Sample 5 Well 8: Sample 6 Well 9: Sample 7 Well 10: Sample 8
mutD gel:
mutL gel:
Transformation of IPTG+GFP+mutL appears to be unsuccessful but samples 4-8 of mutD appear to have the mutD insert as they have higher base pair count.
Quikchange of mutY and DinB was attempted but likely unsuccessful due to lack of any appropriate bands on the below gel and insufficient signal from sequencing
1: Ladder 2: Neg control 3: MutY 4: DinB Quikchange results transformed into XL1Blue cells. Overnight ligation of 10:1 TyrR:promoter(plasmid) attempted. Transformed ParC3, MutY7, DinB1, MutS8, MutH2, MutD5, and MutD8 into XL1Blue Mutation rate assay: Measured OD600 of XL1B wt overnight culture at 12:20: 1.617 (roughly 1.29x10^9 cells/mL) and made an inoculum with 5000 cells/mL in LB (with 1mM IPTG). Made 20 200ul aliquots of inoculum +IPTG and put into incubator at 1:16 pm.
Tuesday
Transformed mCherry+paroF, promoter+TyrR, mCherry+ligase, promoter+ligase, mCherry cut, promoter cut. No growth on mutY and DinB quikchange transformant plates. Attempted 2 more protocols for quikchange for mutY (2-step and 60oC annealing temperature). Both appeared to fail.
1: ladder 2: 60C negative control 3: 60C MutY product 4: 2-Step negative control 5: 2-Step MutY product
2 mutD-positive colonies from 8/18 miniprepped. mutation rate assay: 200ul cultures taken out of 37 degree shaker at 2:30 (~25 hrs), plated (16x) 200 ul on 80ug/mL rif plates as well as 200ul of 10^6 and 10^7 dilutions from 4 different overnight cultures on LB plates. The overnight culture of melA did not grow (the melA plasimid: BBa_K193602 has amp resistance rather than cam as listed on the registry). Inoculated overnight cultures at 5:00 in 5mL LB of XL1B, EmrR3, EmrR4, Dam). Resuspended 2 likely rif resistant colonies and plated on 80ug/mL rif plates to check if rif-resistant bacteria could grow at such a high concentration of rif.
Wednesday
PCRed out and gel-purified TyrR, paroF. Yesterday’s cut plasmids w/o ligase transformants yielded colonies, suggesting that the digestion and dephosphorylaion failed. Re-digested TyrR, paroF, mCherry, promoter and dephosphorylated mCherry and Promoter containing plasmids. Made overnight cultures of yesterday’s mutator transformants (ParC3, MutY7, DinB1, MutS8, MutH2, MutD5, and MutD8) . Redigested Umu’C and mutL and ligated into IPTG+GFP plasmid. melA overnight culture does not look any darker to the naked eye; a melA overnight culture was made with 250ug/mL tyrosine and 1mM IPTG in LB. Mutation rate assay: Dam master inoculum made of ~5000 cfu/mL from yesterday’s overnight culture, appropriate concentration of cam and 1mM IPTG which was then aliquotted to 19 200ul cultures. In addition, a rif80 plate streaked with likely rif-resistant colony from 8/18 have near-lawn growth showing that 80ug/mL rif is not too high a concentration to completely hinder growth. No growth in EmrR3 culture -reinoculated. In addition, XL1Blue overnight cultures were inoculated.
colony counts from XL1B wt test plates (200ul plated of the stated dilution):
| culture | 10^6 dil | 10^7 dil |
|---|---|---|
| 1 | 61 | 8 |
| 2 | 70 | 3 |
| 3 | 63 | 1 |
| 4 | 55 | 3 |
This suggests a cfu/mL concentration of ~5X10^7 was plated on selective plates (a low amount).
Thursday
Overnight cultures of ParC3, MutS, MutH2, DnaE, DinB1, MutY7, MutD5, and MutD8 miniprepped. Made additional overnight cultures of the above for glycerol stocks. mutY quikchange plates from 8/19 show a few colonies on the 2-step mutagenesis samples; these colonies have been used to inoculate overnight cultures. Ran 2-step quikchange protocol on DnaE, ParC, and mutH containing plasmids. Transformed yesterday’s IPTG+GFP+Umu’C/mutL ligation products as well as just IPTG+GFP into XL1Blue cells. paroF and TyrR were PCRed from genomic DNA and fragment isolated. paroF was digested and ligated into the mCherry plasmid, and TyrR into the promoter plasmid and the ligation products transfomed into XL1Blue. Mutation rate assay: 1 rif-resistant colony observed across 16 XL1Blue selective plates (plated on 8/19): likely a function of low plated cfu concentration. Significant growth on 2 rif80 plates platedwith rif-resistant bacteria from 8/19 further confirming that rif concentration is not an issue. Plated yesterday’s Dam 200ul culture aliquots on selective media as well as non-selective test plates (10^6,10^7,10^8 dilutions for three cultures). Made 20 mL culture aliquots of XL1Blue wt and put in 37 degree shaker (up from 200ul). Re-inoculated melA overnights for miniprep tomorrow (melA+tyrosine is not dark/barely grew and we want to sequence).
Friday
Made glycerol stocks of PC3, MS, MH2, DE, DB1, MY7, MD5, and MD8. Quikchange transformant plates from 8/21 show a few small colonies only on quikchanged mutH. Made colony suspensions of yesterday’s Umu’C and mutL ligation product transformants and ran on a gel after colony PCR: Order: Well 1: Ladder Well 2: IPTG+GFP plasmid Well 3: Sample 1 Well 4: Sample 2 Well 5: Sample 3 Well 6: Sample 4 Well 7: Sample 5 Well 8: Sample 6 Well 9: Sample 7 Well 10: Sample 8 MutL gel:
unsuccessful. Umu’C gel:
No amplicons at the correct band (greater than 1kb).
Miniprepped mutY-quikchanged overnight cultures for sequencing. Quikchange transformants from yesterday still show only colonies(5) on the mutH plate. Transformed promoter+TyrR and aroF + mCherry ligation products yielded no colonies so re-ligated and retransformed.
Sunday
Mutation rate assay: no colonies on rif80 plates for Dam (16). Test plates showed inconsistent results (no colonies for culture 1 at any dilution, 1,0,4 colonies for culture 2 at 10^6 10^7, 10^8 dilutions, and 0, 2, 3 colonies for culture 3 at 10^6,10^7,10^8 dilutions). Plated XL1Blue 200ulx18 of the 2ml cultures on rif80 plates as well as 200ul of a 10^6 dilution on agar plates for 3 cultures.
Miniprepped melA but failed to achieve yield over 30ng/ul even with saturated 2x5ml cultures
Week 11 8/25 - 8/29
Monday
We finally got our AroF+mCherry construct ligated in! Ran a colony PCR to verify these, and looks good so far. Made overnight cultures for miniprep tomorrow. We also Dpn1-digested last week’s ParC and DnaE quickchange products and transformed them in.
Par for the course with this, really. We also ran a quickchange with ParC and DnaE again, this time under two different conditions: twice the amount of initial DNA and twice the amount of extension time (which is already too long). We transformed in the 2X DNA samples but the 2X time samples were not transformed until tomorrow. Also ran a colony PCR on last week’s MutL and UmuC ligation attempts: looks like half our UmuC colonies were successful, but all of MutL failed.
Tuesday
Miniprepped the mCherry+AroF constructs while admiring the culture’s pinkish color. Sent that and a UmuC ligation attempt in for sequencing. We also ran another PCR of the TPC and TDE quickchange products (the ones with a doubled extension time), and while one of the TDE samples had promise, none of the TPC samples showed anything. Still went ahead with all the transformations anyway, because these things tend to be wonky at times. We also miniprepped mMutH for sequencing and MelA for verification.
Wednesday
Oh hey, a good sign! Monday’s mDnaE and mParC plates show a possible colony. Made overnight cultures of those and redid our MutS and MutY mutagenesis, with 2X DNA concentrations, since that seemed to have worked well. We transformed those in after Dpn1 digestion. Also sent in mMutH samples for sequencing and religated MutL, MutD, and DinB. We also ligated our promoter+TyrR construct and began a mutation rate assay of wild-type XL1-B cells.
Thursday
Transformed in the constructs we ligated yesterday. Also miniprepped yesterday’s overnight cultures, but not much growth was seen. Miniprep yields were disappointingly low, so there go those samples (i cry evry tym). At least we had colonies grow on our rifampicin plates. Made Dam overnight cultures and new IPTG stock. Friday Really good results for our TyrR construct! Looks like we’re actually starting to make progress! Colony PCR doesn’t seem to be working for this, but hopefully sequencing on Monday will tell us if we got it for certain. No growth on yesterday’s ligation plates, however, but we’re giving the plates one extra day to grow. We seem to be having the same problems that we had a few weeks before. Maybe it’s just a fluke. Also miniprepped yesterday’s overnight cultures. Yields were somewhat low as usual, but looks like we at least have enough for sequencing. Redoing MutS, ParC mutagenesis with a 68C extension temperature.
Saturday
Better results than usual! Apparently we had a colony on mMutY. We still need to get it sequenced, but maybe this one could work? Keeping our fingers crossed. We also miniprepped a bunch of mMutY samples.
Week 12 9/1 - 9/5
Monday--Labor Day
Rikki was the only one who came in on a Federal Holiday. She ran colony PCR on Promoter+TyrR construct (but expected it not to work since for some reason it never amplifies the promoter well--she was right, you couldn’t see a band). She also made overnight cultures of the promoter+TyrR but only made two because she could not find more culture tubes.
Tueday
Miniprepped the two overnight cultures of Promoter+TyrR. We ran out of Promega buffers so we tried invitrogen, but the yields were too poor on one so only one sample was submitted. Guess this was a good thing because--spoiler--the results were negative. We also tried ethanol precipitation for miniprep yields which were too low to submit for sequencing and discovered experimentally that sometimes it works and sometimes it doesn’t. In addition to submitting Promoter+TyrR for sequencing, we also submitted mutagenized MutY and DnaE. DinB, MutD, MutL, UmuC, and IPTG+GFP were digested and the mutators were ligated to IPTG+GFP, this time as an overnight ligation at 4C (Promaga t4 DNA ligase calls for overnights to be done at 4C rather than 16C). We tried site-directed mutagenisis on ParC, MutS, MutY, and DnaE using turbo. Overnight cultures were made of the melanin plasmid, as well as IGDM and IGER (for rif assay).
Wednesday
Transformed ligation products of IPTG+GFP+ DinB, MutD, MutL, UmuC. Assuming failure for Quickchange, trying again but using Turbo, Turbo hot start, Phusion hot start, Q5, and Q5 hot start polymerases side-by-side. Hopefully one will work this time :/ Promoter+TyrR sequencing came back negative today, so 12 overnight cultures were made (since colony PCR doesn’t work well for this construct). Rikki found a random IPTG+GFP+MutD plate in the cold room that looked promising (a lot of colonies, some visibly green, some white). Colony PCR was run on four green colonies and four white colonies.
Thursday
Miniprepped Promoter+TyrR overnights. Rikki meshed together a new protocol using invitrogen buffers but following a lot of Promega’s timing and the yield’s are great--like 500ng/uL). The promoter+TyrR was digested with XbaI and PstI and run on a gel alongside digested promoter:
ladder, promoter, samples 1-5, sample 12
ladder, promoter, samples 6-11
It looks like some of the samples have TyrR!!! (1541 bp) Sample 10 probably didn’t cut right. Submitting samples 1, 3, 12, 7 for sequencing tomorrow morning. Looks promising!!
Colony PCRed MutD:
IPTG+GFP was in the first lane, but must not have been added correctly.We compared the bp size to our IPTG+GFP plasmid and it lookes like 5-8 are just IPTG+GFP (they were picked from green colonies). Samples 1-4 (picked white colonies) appear to have MutD, so they will be miniprepped and sent in for sequencing. Plated EmrR overnight cultures on rif plates and dilutions on control plates (following the rif protocol) and made overnights of EmrR and Dam for the next batch of rif assays. Miniprepped more IPTG+GFP for future ligations.
Friday
Submitted promising Promoter+TyrR and IPTG+GFP+MutD for sequencing. These results should be good!!! Three of the four mutators that were Quikchanged have colonies on the plates! It actually looks like Turbo doesn’t work as well as Q5 of Phusion, which is probably why our results for Quikchange have been pretty bad so far. Overnight cultures were made of these colonies. The ligations of IPTG+GFP+ DinB, MutD, MutL, UmuC don’t appear to have worked, and we’re probably going to scrap anything we don’t get soon.
Week 13 9/8 - 9/12
Summary This was the week that several people (Rikki and Ryan B) were leaving lab and all our stuff suddenly started working last week, so it was an incredibly hectic week where we all stayed in lab until 2 or 3 in the morning every day. Summary of mutator progress by the end of this week (note to Derek/others: we should put this table up each week when available):
| Mutator | Colony PCR? | Seq Verified? | Quikchange? | Overall progress |
|---|---|---|---|---|
| Dam | ✓ | ✓ | Not needed | Complete |
| DinB | X | X | F13V | N/A (may be able to get MIT else retry) |
| DnaE | ✓ | ✓ | E612K | Need sequencing primers to verify mutations! Ordered 9/9 |
| EmrR | ✓ | ✓ | Not needed | Complete |
| MutD | ✓ | ✓ | L73W, A164V | 1st mutation in progress |
| MutH | ✓ | ✓ | E56A | Complete |
| MutL | X | X | E32K | N/A |
| MutS | ✓ | ✓ | G619D | Complete |
| MutT | ✓ | ✓ | Can’t use :( | N/A |
| MutY | ✓ | ✓ | V45A | Need sequencing primers to verify (start from bp 1)! ordered 9/10 |
| ParC | ✓ | ✓ | Y120H | Probably not doing due to the internal cut site. Eliot-Ryan can give it a shot though. |
| UmuC | X (MIT is sending it out. will deal with getting primers for it once we get it 9/11) it requires UmuD’ which has the first 24 amino acids of UmuD removed) | X | Y11A |
Monday
We started PCRs to PCR out the mutators that have been successfully ligated (and Quikchanged when required) from the IPTG-GFP plasmid into the final ParoF-mCherry containing backbone we will actually carry out FREP in. For short, we call this ParoF-mCherry backbone ARM. We also continued various things, such as cloning DinB and MutL into IPTG-GFP, Quikchanging TyrR to make a mutated version of TyrR that has increased sensitivity and range of response to tyrosine, various already ligated things, and inoculating completely mutated Dam, EmrR, alongside wild-type XL1 Blu for the mutation rate assay. To test the promoter response to tyrosine repression, we also inoculated cells transformed with the AroF-TyrR construct into various concentrations of tyrosine (ranging from 0 to around 250 mg/L)
Tuesday
Designed and ordered more oligos that we needed. We continued the Quikchanges and ligations from yesterday. Sequencing results came back, which allowed us to confirm MutS mutagenesis working. However, we’ll probably have to jump ship on ParC unfortunately, because there was an internal cut site that we only just now discovered/bothered to check, which means we only ligated in half of ParC :/ Sadness. We continued trying to PCR out, fragment isolate, and ligate the mutators (Dam EmrR, and MutH) into the ARM construct that we can use for FREP. Measured various OD’s from yesterday’s inoculation for the mutation rate assay, plating a humongous number of plates also (literally hundreds of plates). We also got plate reader data for the assay for promoter response to tyrosine repression, though it looks kinda sketch for one of the strains (appearing to be induced rather than repressed).
Happy birthday Ryan! ^This was at 2 AM on Wednesday morning since that’s how long we were in lab for. Stayed in lab till it was already Ryan’s birthday the next day, what a way to celebrate it XD
When the last people finally left lab at 3AM wednesday morning, we were all incredibly happy as we walked out the door. :D
Wednesday
We needed to make more competent cells (which we inoculated last night), but sadly overgrew it, so we’re gonna reinoculate and try again tomorrow. We transformed ARM ligations from yesterday, PCR’d to get linearized amp backbone we’ll need for the final functional TyrR construct (right now it’s on a chloramp backbone, but we need it on amp since the mutator construct will already be on chloramp). We ran colony PCR on the MutL and DinB ligations, both of which luckily showed lots of colonies.
Thursday
Sad farewells :( Rikki and Ryan B left last night, so we are down to two people manning the lab. Eliot will be back next week and Kevin will be leaving after Friday. We miniprepped MutS and mutated TyrR (the latter of which needs to be sent in for sequencing verification of the mutagenesis) from yesterday. We also collected data for the mutation rate assay:
| E | XLB | Dam | |
|---|---|---|---|
| 1 | 0 colonies | 5 colonies | 0 colonies |
| 2 | Near-lawn to lawn growth. Bad plate? | 4 colonies | 0 colonies |
| 3 | 0 colonies | 7 colonies | 0 colonies |
| 4 | 0 colonies | 0 colonies | 0 colonies |
| 5 | 0 colonies | 4 colonies | 0 colonies |
| 6 | 0 colonies | 6 colonies | 0 colonies |
| 7 | 0 colonies | 10 colonies | 0 colonies |
| 8 | 0 colonies | 1 colonies | 0 colonies |
| 9 | N/A | 8 colonies | 0 colonies |
| 10 | N/A | 2 colonies | 0 colonies |
| 11 | N/A | 11 colonies | 1 colonies |
| 12 | N/A | 10 colonies | 0 colonies |
| 13 | N/A | 0 colonies | 0 colonies |
| 14 | N/A | N/A | Bad plate? 1 cluster of colonies |
| 15 | N/A | N/A | 0 colonies |
| 16 | N/A | N/A | 0 colonies |
Looks like most of the mutator expressing plates have few colonies compared to wild type for whatever reason. Did colony PCR on the ARM and IPTG-GFP ligations from previous days, and found a few suitable colonies to send in (MutL specifically), though we need to do more colony PCR on some of the ligations (DinB, EmrR and DnaE specifically). We also have been collecting lysate from cells transformed with the various mutators we’ve been ligating, and ran it on an SDS-PAGE gel to confirm that the IPTG-GFP plasmids are properly expressing the mutators. We have a pic of the gel on the next day. Ladder
- XLIB/IPTG (9/9) (7)
- XLIB+IPTG (9/9) (1)
- XLIB+IPTG (9/9) (5)
- XLIB+IPTG (9/9) (6)
- XLIB+IPTG (9/4)
- EmrR+IPTG (9/9) (8)
- EmrR+IPTG (9/9) (10)
- EmrR+IPTG (9/4) (1)
- EmrR+IPTG (9/4) (2)
- EmrR+IPTG (9/4) (3)
- EmrR/IPTG (9/9) (3)
- EmrR/IPTG (9/9) (4)
- EmrR/IPTG (9/4) (1)
- EmrR/IPTG (9/4) (2)
- Dam+IPTG (9/9) (9)
- Dam/IPTG (9/9) (2)
- Dam/IPTG (9/9) (11)
Friday
We somehow lost our old mutated DnaE samples, so we went through the process of re-Quikchanging wild-type DnaE (alongside our newly ligated MutL). We also plan to ligate our sequence-verified, mutagenized MutS into ARM, so we set up the digests for that today, will need to ligate tomorrow. We ran more Colony PCRs of the ones that didn’t show any positives from before (see Thursday), and finally got more positives for EmrR and DamR, but still not for DinB (likely we’ll have to . We finished processing/imaging the protein gel we ran yesterday. A link here File:UChicago wk13gel1.tif. Note: the protein gel is inverted so lane labels as listed from yesterday are backwards. The two rows should be identical; they’re just done at different protein concentrations to see which one would image better. We got some plasmids from Walker lab at MIT also today, which we put on nonselective LB plates for recovery. We may need these plasmids for PolB and UmuD’C, but on second thoughts (as you’ll see later), we ended up not actually needing them. We checked Dam and EmrR plates from the mutation rate assay that were plated on Thursday for growth, but there were few/no colonies, so we’ll wait one more day and see what happens as time works its magic.
Saturday
Ryan S came in for a bit today to do various household chores. Checked plates we restreaked yesterday from glycerol stock to repeat the mutation rate assay, restreaked Walker lab plasmids from yesterday on selective plates (cam for the PolB plasmid and amp for the UmuD’C plasmid). He checked Dam and EmrR plates from the mutation rate assay that were plated on Thursday once again for growth and recorded down counts. Transformed various Quikchanges and ligations from earlier in the week we had been stalling an transforming (we wanted to transform a whole bunch of things at once to make things more efficient). Finally, made overnight cultures of the colony PCRs from Friday that looked good for EmrR and DamR that had been ligated into the ARM construct.
Week 14 9/15 - 9/19
Summary
Running out of time/last final stretch: Everything’s coming together, but we still have a ton of work to do.
Sunday
Ryan S counted colonies on the nonselective XL1B plates and checked all the plates from yesterday. None of the transformation have large colonies so they were put for an additional day. Miniprepped the overnight cultures of Dam and EmrR that actually grew. We have one of each! UmuD’C and PolB from the selective plates were innoculated and cultured in selective media. Made glycerol stocks of the 4 mutators we have, and performed mutation rate analysis. Made a mastermix in LB, rif and IPTG at the right cell dilution and aliquotted out for overnight cultures. They’re now in the incubator and ready for tomorrow’s plating!
Monday
Checked all the transformation plates but none of them seems to have growth :( Good news is, the overnight cultures of the mutators for mutation rate analysis (EmrR, MutS, MutH, Dam) have growth, so they were plated on LB plates and Rif plates when the right OD is reached. DinB and UmuD’C were digested and ligated onto IPTG+GFP. PolB and UmuD’C were miniprepped and glycerol stocks were made. The miniprep sample concentration was really high - they were around 500 ng/ul!
Tuesday
UmuD’C and linearized amp backbone. Digested the UmuD’C, DinB and Dam and ligated them into IPTG+GFP with a 3:1 vector-insert ratio. AREmrR was sent in for sequencing and overnight cultures for ARDam was made for miniprepping. Transformation plates from yesterday and the day before looked bad. The nonselective plates showed a lot of growth while the selective plates don’t. Seems like we need to do ligations and quickchange again…
Eliot then worked frantically to get something done! He quikchanged MutD, DnaE, MutL and TyrR. Then, he re-ligated UmuD’C and DinB into IPTG+GFP. Sequencing results: We have our mutated samples of mMutL, mMutY and mMutS and AroF+mCherry+mMutH! There was an extraneous mutation in TyrR that we used for all the ligations though. Fortunately, one of the templates (PT3) has a silent mutation. We could continue using this and make new quikchange templates of TyrR.
Wednesday
Sent in ARMEmrR and ARMDam for sequencing! However, overnights of ARMDam appear to have no growth...Made overnights again for miniprep and glycerol stocks. WE FOUND A GLYCEROL STOCK FOR THE ARM BACKBONE!! Quickly plated them for miniprep.
Thursday
ARMDam overnights finally have growth! Glycerol stocks made and miniprepped. Checked the mutation rate assay plates but not all of the selective plates have growth. We’ll check them again tomorrow! Made overnight cultures of XL1-B and JW-1316-1 for competent cells and ARM backbone for miniprepping. Also digested and ligated mMutY and mMutS into ARM backbone with 5:1 ratio. We ran out of competent cells so we’ll transform the products once the competent cells are made!
Friday
Miniprepped ARM backbone from yesterday’s overnight cultures. XL1-B and JW-1316-1 were cultured and competent cells were made. What a long day in the cold room! The cells were tested for their transformation efficiency using the iGEM kit. Quikchanged MutL, MutD, DnaE and TyrR. Ligations from yesterday and these quikchanged products were transformed into competent Xl1-B and plated.
Saturday
Transformation efficiency of our competent cells is approximately 2X10^6. Hmmm it’s a bit lower than what we want but we’ll just go with it. More overnight cultures of the mutators on IPTG+GFP were made for the mutation rate analysis!
Week 15 9/22 - 9/26
Summary
It’s the final countdown! And we still don’t (and probably won’t) have the final FREP stuff good to go! (*Panicking intensifies*). But don’t FRET over FREP! McGuire and Eliot will do their best to finish what they can and lament what they cannot.
Sunday
Ryan continued the Mutation Rate Assay protocol, checking ODs and making according overnight cultures. He also autoclaved various items and transformed Q1-5, P1-5, ARMY, ARMS, and ARMS1 for Eliot.
Monday
Ryan checked ODs and plated cultures for the Mutation Rate Assay. He also started the final Mutation Rate Assay attempt, using XL1B with IPTG+GFP, Dam, IGER4, mMutH1, and mMutH2. McGuire also autoclaved a small boatload of micropipette tips. All work on cloning/mutagenesis was stopped today. End results:
| Mutator | Colony PCR? | Seq Verified? | Quikchange? | Overall progress |
|---|---|---|---|---|
| Dam | ✓ | ✓ | Not needed | Complete |
| EmrR | ✓ | ✓ | Not needed | Complete |
| MutH | ✓ | ✓ | E56A | Complete |
| MutY | ✓ | ✓ | V45A | Complete |
Eliot started concentrating on the tyrosine assay. Since ligating TyrR into ARM would take too long and be subject to setbacks, he used only the ARM backbone with endogenous, wild-type TyrR in DH5a. Eliot transformed TyrR into DH5A cells today, growth in LB with TyrR to come on Tuesday.
Tuesday
Ryan continued the Mutation Rate Assay protocol, checking ODs and making according overnight cultures. Also checked the nonselective plates from Monday. Eliot delayed transformation of the ARM backbone delayed since either he goofed and added DNA to the negative control or contamination or plates were bad or something. Ergo, Eliot decided to redo the transformation, making sure to grab a plate from a different batch this time.
Wednesday
Ryan checked ODs and plated cultures for the Mutation Rate Assay and made 40 more rif plates. Eliot’s transformation went much better this time. Negative control looked weird: there might be some growth but the agar in this plate was weird, so it could have been the agar looking like that. ARM plate actually showed good growth, however, compared to the other plate. Eliot made the tyrosine cultures and added samples of 3 colonies to each. Needed to match the tyrosine concentrations, though. Eugene suggested starting from a controlled concentration and diluting it down, so trying that. Planning to pick 3 colonies, add them to different concentrations of tyrosine. Concentrations: 0-500mg/L, in 50mg/L intervals (50, 100, 150…) 3 colonies picked, suspended in 35uL H2O each. 2uL suspended colonies put in 5uL LB/Tyr
Thursday
Eliot took cultures at 12:05 PM, total incubation period is 16 hours. Successful growth occurred in most cultures, with the exception of 2 vials: should not affect results too much if data is consistent across all 3 colonies, as the vials correspond to different colonies and are not in the range of 200-300mg/L tyrosine, which appears to be the point at which AroF is repressed judging from the previous assay. He loaded approx. 200 µL culture into wells of 96-well plate. Rows A, B, and C corresponded to colonies 1, 2, and 3. Columns 1-12 were: 0=LB, no cells, 1, 0mg/uL tyrosine, 2=50mg/L tyr, 3= 100mg/L tyr, … 12= 500mg/L tyr. Measured 570-610mm. Ryan checked nonselective plates for the Mutation Rate Assay. All fluoresced (yay)! Friday Ryan and Eliot were both busy, so no one went into the lab on this day of Fries.
Saturday
Ryan stared across the barren wasteland lab, wondering if it was all worth it. Had all the blood, sweat, and tears been shed for this? This...end? It seemed that this experiment shall go out with a whimper rather than a bang...Ah, well. Ryan checked the selective plates for the Mutation Rate Assay. All of these had fluorescing colonies as well. McGuire headed to the Museum of Science and Industry to drink some blue lemonade for free whilst standing inside of a manmade mini-tornado.
 "
"