Team:Bordeaux/Results
From 2014.igem.org
m |
m |
||
| Line 4: | Line 4: | ||
[[file:Bdx2014 Production.jpg|left|400px]] [https://2014.igem.org/Team:Bordeaux/Results/Production Production (click for more info)]<br><br><br> We managed to settle the best conditions to produce the biopolymers in maximal amount. Different medium and production time were tested and a special BL21 strain was used.<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | [[file:Bdx2014 Production.jpg|left|400px]] [https://2014.igem.org/Team:Bordeaux/Results/Production Production (click for more info)]<br><br><br> We managed to settle the best conditions to produce the biopolymers in maximal amount. Different medium and production time were tested and a special BL21 strain was used.<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
---- | ---- | ||
| - | [[File:Bdx2014 Purification.jpg|left|400px]] [https://2014.igem.org/Team:Bordeaux/Results/Purification Purification (click for more info)]<br><br><br> The special properties of the ELP | + | [[File:Bdx2014 Purification.jpg|left|400px]] [https://2014.igem.org/Team:Bordeaux/Results/Purification Purification (click for more info)]<br><br><br> The special properties of the ELP were used to purify the produced proteins by inverse thermal cycling (ITC), which is a quick, easy and column-free method for purification of this polymer <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
---- | ---- | ||
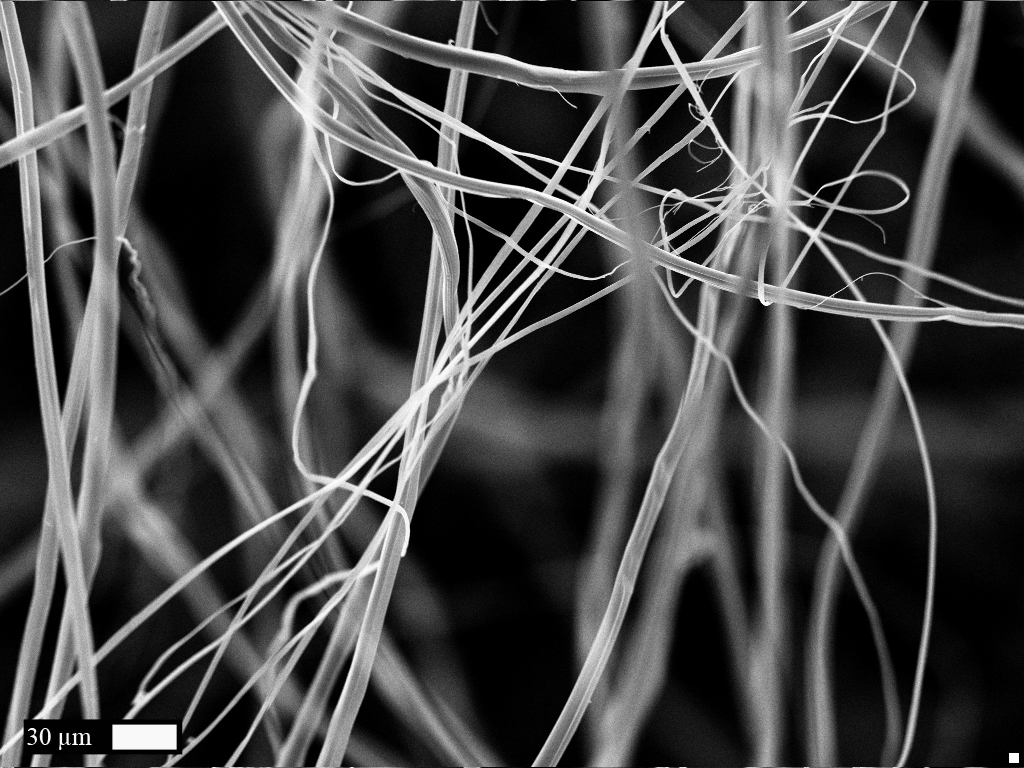
| - | [[File:Bdx2014 Characterization.jpg|left|400px]] [https://2014.igem.org/Team:Bordeaux/Results/Characterization Characterization (click for more info)]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | + | [[File:Bdx2014 Characterization.jpg|left|400px]] [https://2014.igem.org/Team:Bordeaux/Results/Characterization Characterization (click for more info)]<br><br><br> Characterization was performed according to different physical and chemical methods. As the protein produced is a polymer, it was transformed into a fiber and its resistance and elasticity have been tested. <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
Latest revision as of 02:20, 18 October 2014



We managed to settle the best conditions to produce the biopolymers in maximal amount. Different medium and production time were tested and a special BL21 strain was used.
Purification (click for more info)
The special properties of the ELP were used to purify the produced proteins by inverse thermal cycling (ITC), which is a quick, easy and column-free method for purification of this polymer
Characterization (click for more info)
Characterization was performed according to different physical and chemical methods. As the protein produced is a polymer, it was transformed into a fiber and its resistance and elasticity have been tested.
References
[1] Doreen M. Floss et al. ELASTIN-like polypeptides revolutionize recombinant protein expression and their biomedical application. Trends in Biotechnology Vol.28 No.1 (PMID 19897265)
[2] Dan W. Urry Entropic Elastic Processes in Protein Mechanisms. I. Elastic Structure Due to an Inverse Temperature Transition and Elasticity Due to Internal Chain Dynamics. Journal of Protein Chemistry, Vol. 7, No.. I, 1988
[3] Dan W. Urry. Physical Chemistry of Biological Free Energy Transduction As Demonstrated by Elastic Protein-Based Polymers. J. Phys. Chem. B 1997, 101, 11007-11028
[4] Dan E. Meyer and Ashutosh Chilkoti. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. NATURE BIOTECHNOLOGY VOL 17 NOVEMBER 1999
[5] Trabbic-Carlson et al. (2004), Expression and purification of recombinant proteins from Escherichia coli: Comparison of an elastin-like polypeptide fusion with an oligohistidine fusion. Protein Science, 13: 3274–3284. doi: 10.1110/ps.04931604
[6] K. Trabbic‐Carlson et al. Effect of protein fusion on the transition temperature of an environmentally responsive elastin‐like polypeptide: a role for surface hydrophobicity? Protein Engineering, Design and Selection (2004) 17 (1): 57-66. doi: 10.1093/protein/gzh006
 "
"