Team:Austin Texas/photocage
From 2014.igem.org
Jordanmonk (Talk | contribs) |
|||
| (4 intermediate revisions not shown) | |||
| Line 125: | Line 125: | ||
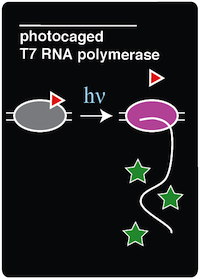
After this exposure, the cells were grown for an additional 16 hours, allowing the newly decaged T7 RNAP to transcribe the GFP reporter gene, which ultimately results in fluorescence. As can be seen in '''Figure 3''', while the fluorescence background at time zero is not as low as we would like, there is a clear and dramatic increase in fluorescence that is directly dependent on the amount of time the culture was exposed to 365 nm light. This result is consistent with previous results using this system (Chou et al. 2010). | After this exposure, the cells were grown for an additional 16 hours, allowing the newly decaged T7 RNAP to transcribe the GFP reporter gene, which ultimately results in fluorescence. As can be seen in '''Figure 3''', while the fluorescence background at time zero is not as low as we would like, there is a clear and dramatic increase in fluorescence that is directly dependent on the amount of time the culture was exposed to 365 nm light. This result is consistent with previous results using this system (Chou et al. 2010). | ||
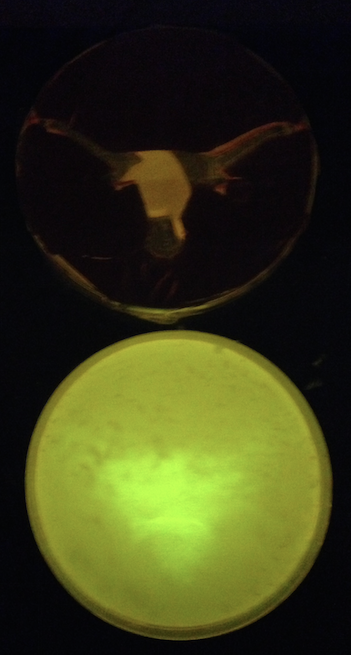
| - | + | We also tested the spatial activation of T7 RNAP activity by placing a cutout above a petri dish and shining 365 nm light through it ('''Figure 4'''). Although cells were present uniformly across the entire surface, we only observed GFP expression in the areas exposed to light, as expected. | |
| Line 138: | Line 138: | ||
<h1>Discussion</h1> | <h1>Discussion</h1> | ||
| - | [[File:Longhorn photocage GFP.png|200px|thumb|right|Photocage culture plated onto ONBY solid media and exposed to | + | [[File:Longhorn photocage GFP.png|200px|thumb|right|'''Figure 4.''' Photocage culture plated onto ONBY solid media and exposed to 365 nm light under a UT Longhorn stencil for 15 minutes. Although not yet perfected, the system is indeed light-activated and shows promise.]] |
The data from our photocage project provides strong evidence that we have, in fact, replicated the light-activated protein expression system that Chou et al. had constructed. | The data from our photocage project provides strong evidence that we have, in fact, replicated the light-activated protein expression system that Chou et al. had constructed. | ||
| Line 152: | Line 152: | ||
<h1>References</h1> | <h1>References</h1> | ||
| - | *Chou, C., Young, D. D. and Deiters, A. (2010), Photocaged T7 RNA | + | *Achenbach, J. C., Chiuman, W., Cruz, R. P. G., Li, Y. (2004), DNAzymes: from creation in vitro to application in vivo.''Curr. Pharm. Biotechnol.'' '''5''':321 –336. |
| - | *Deiters, A., Groff, D., Ryu, Y., Xie, J. and Schultz, P. G. (2006), A | + | *Chou, C., Young, D. D. and Deiters, A. (2010), Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. ''ChemBioChem'' '''11''':972–977. |
| - | *P. A. | + | *Deiters, A., Groff, D., Ryu, Y., Xie, J. and Schultz, P. G. (2006), A genetically encoded photocaged tyrosine. ''Angew. Chem.'' '''118''':2794–2797. |
| - | *D. | + | *Osumidavis, P. A., Sreerama, N., Volkin, D. B., Middaugh, C. R., Woody, R. W., Woody, A. Y. M. (1994), Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. ''J. Mol. Biol.'' '''237''':5–19. |
| - | + | *Temiakov, D., Patlan, V., Anikin, M., McAllister, W. T., Yokoyama, S., Vassylyev, D. G.. (2004), Structural basis for substrate selection by T7 RNA polymerase. ''Cell'' '''116''':381–391. | |
| - | + | ||
</div> | </div> | ||
<!-- WIKI CONTENT ENDS --> | <!-- WIKI CONTENT ENDS --> | ||
Latest revision as of 03:03, 18 October 2014
| |||||||||||||||||||||||||||||
 "
"