Team:Heidelberg/pages/Collaborations
From 2014.igem.org
| (16 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
=iGEM Team Aachen= | =iGEM Team Aachen= | ||
| - | |||
Michael from the iGEM team Aachen visited us in Heidelberg. Their team worked on the development of a real-time pathogen detection technique for what they have build their own fluorescence measurement camera. We provided our expression vectors to the iGEM team Aachen, which they used to express their part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1319003 E1010-K1319003], the human galectin-3 protein fused to mRFP. Nicely seen in Figure 1 and 2 the the intensity of the red color is significantly higher for the induced sample with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362091 pSBX1A30] compared to the uninduced ones. To ensure the right molecular mass of the fusion protein, a SDS gel was run as well (Figure 3). | Michael from the iGEM team Aachen visited us in Heidelberg. Their team worked on the development of a real-time pathogen detection technique for what they have build their own fluorescence measurement camera. We provided our expression vectors to the iGEM team Aachen, which they used to express their part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1319003 E1010-K1319003], the human galectin-3 protein fused to mRFP. Nicely seen in Figure 1 and 2 the the intensity of the red color is significantly higher for the induced sample with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362091 pSBX1A30] compared to the uninduced ones. To ensure the right molecular mass of the fusion protein, a SDS gel was run as well (Figure 3). | ||
| Line 25: | Line 24: | ||
=iGEM Team Tuebingen= | =iGEM Team Tuebingen= | ||
| - | The project of iGEM Team Tuebingen is about blood types and the difficulties that arises during blood donation of the wrong blood type. They aim to develop an easy system to convert blood types A, B, AB to the rares but also most applicable blood type 0. They have applied the method of intein targeting, a method which can also be found in our intein toolbox, to the N-Acetyl-Galactosaminidase protein. More about their project can be read in the very nice [http://igem14-heidelberg.tumblr.com/post/98138990630/igem-team-tubingen article] they have wrote for our tumblr blog or on their own [Team:Tuebingen wiki]. | + | The project of iGEM Team Tuebingen is about blood types and the difficulties that arises during blood donation of the wrong blood type. They aim to develop an easy system to convert blood types A, B, AB to the rares but also most applicable blood type 0. They have applied the method of intein targeting, a method which can also be found in our intein toolbox, to the N-Acetyl-Galactosaminidase protein. More about their project can be read in the very nice [http://igem14-heidelberg.tumblr.com/post/98138990630/igem-team-tubingen article] they have wrote for our tumblr blog or on their own [[Team:Tuebingen|wiki]]. |
| - | In this collaboration, Tuebingen conducted a mass spectrometry analysis of our Lambda lysozyme to proof | + | In this collaboration, Tuebingen conducted a mass spectrometry analysis of our Lambda lysozyme to proof circularization. The data still has to be evaluated, but we will have them ready until the jamboree. There is a high demand for our expression vectors. Tuebingen had problems with expressing their constructs, therefore we send them some of our plasmids pSBX1K30 ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362093 BBa_K1362093]) and pSBX4K50 ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362097 BBa_K1362097]), as they showed to work really nicely even with big proteins like the [[Team:Heidelberg/Project/PCR2.0|Dnmt1]] with 105kDa. |
| - | + | <br/> | |
| + | =iGEM Team Freiburg= | ||
{{#tag:html| | {{#tag:html| | ||
| Line 57: | Line 57: | ||
<div class="carousel-inner"> | <div class="carousel-inner"> | ||
<div class="item active"> | <div class="item active"> | ||
| - | + | <a class="img-enlarge " href="https://static.igem.org/mediawiki/2014/c/c9/Heidelberg_orig_Freiburgvonoben2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2014/c/c9/Heidelberg_orig_Freiburgvonoben2.jpg" class="img-responsive" alt="Image"></a> | |
</div> | </div> | ||
<div class="item"> | <div class="item"> | ||
| - | + | <a class="img-enlarge" href="https://static.igem.org/mediawiki/2014/2/20/Heidelberg_orig_Freiburg_presentation2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2014/2/20/Heidelberg_orig_Freiburg_presentation2.jpg" class="img-responsive" alt="Image"></a> | |
</div> | </div> | ||
<div class="item"> | <div class="item"> | ||
| - | + | <a class="img-enlarge" href="https://static.igem.org/mediawiki/2014/c/c6/Heidelberg_orig_Freiburg_Inductionbox2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2014/c/c6/Heidelberg_orig_Freiburg_Inductionbox2.jpg" alt="Image"></a> | |
</div> | </div> | ||
| - | + | <div class="item"> | |
| - | + | <a class="img-enlarge" href="https://static.igem.org/mediawiki/2014/d/d1/Heidelberg_orig_Freiburg_Cellculture2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2014/d/d1/Heidelberg_orig_Freiburg_Cellculture2.jpg" class="img-responsive" alt="Image"></a> | |
</div> | </div> | ||
| - | + | <div class="item"> | |
| - | + | <a class="img-enlarge" href="https://static.igem.org/mediawiki/2014/7/7c/Heidelberg_orig_Freiburg_discussion2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2014/7/7c/Heidelberg_orig_Freiburg_discussion2.jpg" class="img-responsive" alt="Image"></a> | |
</div> | </div> | ||
| - | + | <div class="item"> | |
| - | + | <a class="img-enlarge" href="https://static.igem.org/mediawiki/2014/8/88/Heidelberg_orig_Freiburg_Teamfoto2.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2014/8/88/Heidelberg_orig_Freiburg_Teamfoto2.jpg" class="img-responsive" alt="Image"></a> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 103: | Line 91: | ||
After cooperation with the iGEM Team Freiburg for two years in a row we are hoping to keep Freiburg as a reliable partner in the next years as well. | After cooperation with the iGEM Team Freiburg for two years in a row we are hoping to keep Freiburg as a reliable partner in the next years as well. | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | =iGEM Team Marburg= | ||
| + | |||
| + | The screensaver of [[Team:Heidelberg/Human_Practice/igemathome|iGEM@home]] is a great possibility to spread news and information about Synthetic Biology around the world. Lately, even other iGEM team have used this new option and filled our screensaver with slides about their own project. Marburg was able to promote their quiz app about Synthetic Biology. | ||
| + | |||
| + | <html> | ||
| + | <style> | ||
| + | #propose {text-indent: 40px; | ||
| + | } | ||
| + | #greetings {text-indent: 20px; | ||
| + | } | ||
| + | ul.supporter_team_list { list-style-type: none; | ||
| + | } | ||
| + | ul.supporter_team_list a {text-decoration: none;color: black;} | ||
| + | ul.supporter_team_list a:hover {text-decoration: underline;} | ||
| + | ul.supporter_team_list > li {display:inline;margin: 30px; | ||
| + | } | ||
| + | </style> | ||
| + | <div id="socioeconomics"> | ||
| + | <h1>A proposal of the German iGEM Teams concerning Intellectual Property</h1> | ||
| + | <p>During the meetup of the German iGEM teams from 23rd to 25th May also <a href="https://2014.igem.org/Meetups:May_LMU-Munich/Workshops">workshops</a> took place in which amongst others we discussed the topic of bioethics. Moral questions were addressed, regarding the value of life and human influence on it, as well as questions dealing with the possible socioeconomic effects of synthetic biology. | ||
| + | </p> | ||
| + | <p> | ||
| + | Especially the topic of an open source vs. patent controlled field accounted for a large part of the discussion. During the discussion one student brought up the point that the legal status of parts in registry remains unclear and that there are parts (e.g. <a href="http://parts.igem.org/Part:BBa_K180009">BBa_K180009</a>) where only upon a closer look it becomes clear that the rights are company–owned. The issue that the legal status of parts in the registry remains uncertain is also mentioned in a recent article published by Nature (<a href="http://www.nature.com/news/synthetic-biology-cultural-divide-1.15149">Bryn Nelson ‘Synthetic Biology: Cultural Divide ’, Nature 509, 152–154, 08 May 2014</a>) : | ||
| + | </p> | ||
| + | <p> | ||
| + | <i>"[N]o one can say with any certainty how many of these parts are themselves entirely free of patent claims."</i> | ||
| + | </p> | ||
| + | <p> | ||
| + | We, the German iGEM teams, therefore like to suggest the addition of a new feature to the parts registry: | ||
| + | </p> | ||
| + | <p id="propose"> A dedicated data field of license information for each BioBrick part.</p> | ||
| + | <p> | ||
| + | For the implementation, we propose to introduce two new fields to BioBrick part entries in the registry: | ||
| + | </p> | ||
| + | <ol> | ||
| + | <li>A string property "LicenseInfo"</li> | ||
| + | <li>A traffic light property (grey, green, yellow, red) to indicate the level of legal protection (unknown, BPA-like, free for research purposes, heavily protected)</li> | ||
| + | </ol> | ||
| + | |||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/6/69/TU-BS_Part_licence_info.png" alt="This image shows a how the proposed field LicenceInfo could look like in the registry."> | ||
| + | |||
| + | <p> | ||
| + | Implementing this feature would in our opinion further clarify and extend the parts info, provide a machine-readable format and thus improve future entries. With the emerging Entrepreneurship track and applications getting closer to industrial realization, the legal status becomes more and more important. Also it would raise awareness to the topic of the legal status of parts, leading to a debate which could further promote the idea of open source. At the same time we hope that examination of most parts will show that they are indeed free of restrictive legal protections. | ||
| + | </p> | ||
| + | <p id="greetings"> | ||
| + | The German iGEM Teams, | ||
| + | </p> | ||
| + | <ul class="supporter_team_list"> | ||
| + | <li><a href="https://2014.igem.org/Team:Aachen">Aachen </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Berlin">Berlin </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Bielefeld-CeBiTec">Bielefeld-CeBiTec </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Braunschweig">Braunschweig </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Freiburg">Freiburg </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Goettingen">Goettingen </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Hannover">Hannover </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Heidelberg">Heidelberg </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:LMU-Munich">LMU-Munich </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Marburg">Marburg </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Saarland">Saarland </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:Tuebingen">Tuebingen </a></li> | ||
| + | <li><a href="https://2014.igem.org/Team:TU_Darmstadt">TU Darmstadt </a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 03:54, 18 October 2014
Besides spending a whole summer in the lab and developing an own project, iGEM is about meeting new people, people with the same interests, the same motivation, the same dedication. Collaborations between team ensure the exchange of ideas, therefore helping the field of Synthetic Biology to be interdisciplinary and creative - the best conditions to give rise to revolutionary projects. We are very happy to met some of the team members from Freiburg, Aachen, Tuebingen, Marburg and London.
Contents |
iGEM Team Aachen
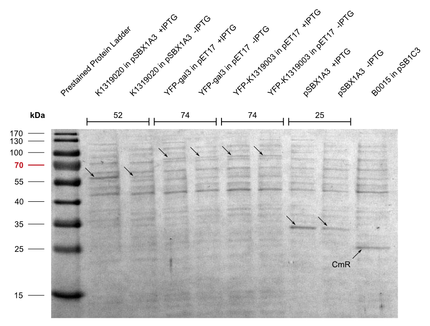
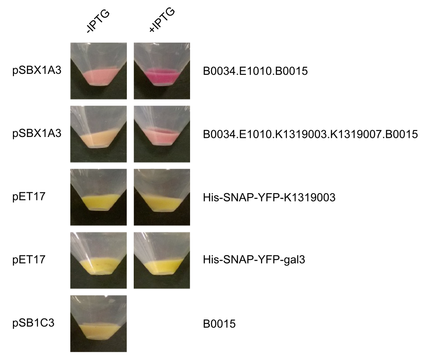
Michael from the iGEM team Aachen visited us in Heidelberg. Their team worked on the development of a real-time pathogen detection technique for what they have build their own fluorescence measurement camera. We provided our expression vectors to the iGEM team Aachen, which they used to express their part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1319003 E1010-K1319003], the human galectin-3 protein fused to mRFP. Nicely seen in Figure 1 and 2 the the intensity of the red color is significantly higher for the induced sample with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362091 pSBX1A30] compared to the uninduced ones. To ensure the right molecular mass of the fusion protein, a SDS gel was run as well (Figure 3).
More information about their project can be found on Aachens Galectin-3 site.
In exchange they offered to characterise these expression vectors with their own designed analyser by measuring the amount of produced fluorescent proteins. Unfortunately, transporting E.colis to another city wasn't as easy as expected, not all Bacteria were able to grow afterwards and we could only obtain sufficient data from pSBX4C5.
iGEM Team Tuebingen
The project of iGEM Team Tuebingen is about blood types and the difficulties that arises during blood donation of the wrong blood type. They aim to develop an easy system to convert blood types A, B, AB to the rares but also most applicable blood type 0. They have applied the method of intein targeting, a method which can also be found in our intein toolbox, to the N-Acetyl-Galactosaminidase protein. More about their project can be read in the very nice [http://igem14-heidelberg.tumblr.com/post/98138990630/igem-team-tubingen article] they have wrote for our tumblr blog or on their own wiki.
In this collaboration, Tuebingen conducted a mass spectrometry analysis of our Lambda lysozyme to proof circularization. The data still has to be evaluated, but we will have them ready until the jamboree. There is a high demand for our expression vectors. Tuebingen had problems with expressing their constructs, therefore we send them some of our plasmids pSBX1K30 ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362093 BBa_K1362093]) and pSBX4K50 ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362097 BBa_K1362097]), as they showed to work really nicely even with big proteins like the Dnmt1 with 105kDa.
iGEM Team Freiburg
Three of our team members, Anna, Caro, and Charlotte visited the iGEM team Freiburg. We met the team right after a big team meeting, therefore we were able to hear their latest project developments. We spend the evening talking about experiences in the lab and about iGEM. It was relieving to hear that they faced similar difficulties in the lab. The morning after we presented our project to the iGEM team Freiburg. Freiburg was very interested in our program iGEM@home as we offered them to promote their own human practice projects. You can also find an [http://igem14-heidelberg.tumblr.com/post/99936805780/optogenetics-the-lock-and-key-for-daily-life article] about optogenetics on our tumblr blog. Both our teams are working with light inducible systems, manly the LOV domain. Although working in different organisms, the constructs were compatible, therefore we could interchange protocols for induction and preliminary results. Furthermore we gazed at their professional light induction box - we can definitely improved ours.
After cooperation with the iGEM Team Freiburg for two years in a row we are hoping to keep Freiburg as a reliable partner in the next years as well.
iGEM Team Marburg
The screensaver of iGEM@home is a great possibility to spread news and information about Synthetic Biology around the world. Lately, even other iGEM team have used this new option and filled our screensaver with slides about their own project. Marburg was able to promote their quiz app about Synthetic Biology.
A proposal of the German iGEM Teams concerning Intellectual Property
During the meetup of the German iGEM teams from 23rd to 25th May also workshops took place in which amongst others we discussed the topic of bioethics. Moral questions were addressed, regarding the value of life and human influence on it, as well as questions dealing with the possible socioeconomic effects of synthetic biology.
Especially the topic of an open source vs. patent controlled field accounted for a large part of the discussion. During the discussion one student brought up the point that the legal status of parts in registry remains unclear and that there are parts (e.g. BBa_K180009) where only upon a closer look it becomes clear that the rights are company–owned. The issue that the legal status of parts in the registry remains uncertain is also mentioned in a recent article published by Nature (Bryn Nelson ‘Synthetic Biology: Cultural Divide ’, Nature 509, 152–154, 08 May 2014) :
"[N]o one can say with any certainty how many of these parts are themselves entirely free of patent claims."
We, the German iGEM teams, therefore like to suggest the addition of a new feature to the parts registry:
A dedicated data field of license information for each BioBrick part.
For the implementation, we propose to introduce two new fields to BioBrick part entries in the registry:
- A string property "LicenseInfo"
- A traffic light property (grey, green, yellow, red) to indicate the level of legal protection (unknown, BPA-like, free for research purposes, heavily protected)

Implementing this feature would in our opinion further clarify and extend the parts info, provide a machine-readable format and thus improve future entries. With the emerging Entrepreneurship track and applications getting closer to industrial realization, the legal status becomes more and more important. Also it would raise awareness to the topic of the legal status of parts, leading to a debate which could further promote the idea of open source. At the same time we hope that examination of most parts will show that they are indeed free of restrictive legal protections.
The German iGEM Teams,
 "
"