Team:Marburg:Project:Silver
From 2014.igem.org
| (22 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
SilverSURF | SilverSURF | ||
{{Team:Marburg/Template:StartContinue}} | {{Team:Marburg/Template:StartContinue}} | ||
| + | {{Team:Marburg/Template:Submenu:Project:Silver}} | ||
| + | <html><a name="intro"><h2>Introduction</h2></a></html> | ||
"Surfing is a surface water sport in which the wave rider, referred to as a surfer, | "Surfing is a surface water sport in which the wave rider, referred to as a surfer, | ||
| Line 15: | Line 17: | ||
increasing signals for detection. Here we use the bacterial flagellum, a structure provided by nature itself, | increasing signals for detection. Here we use the bacterial flagellum, a structure provided by nature itself, | ||
as a toolbox for different applications. This huge formation consists | as a toolbox for different applications. This huge formation consists | ||
| - | of the basal body, the hook and the filament, which is composed of up to 20.000 copies of the protein Flagellin (Figure 1). Due to the rotating flagellum bacteria are able to | + | of the basal body, the hook and the filament, which is composed of up to 20.000 copies of the protein Flagellin (Figure 1). Due to the rotating flagellum, bacteria are able to |
| - | surf through their environment. | + | "surf" through their environment. |
<html> | <html> | ||
| Line 22: | Line 24: | ||
<img src="https://static.igem.org/mediawiki/2014/4/49/MR_project_1.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/4/49/MR_project_1.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 1: Composition of the bacterial | + | <b>Figure 1: Composition of the bacterial flagellar filament.</b> |
</span> | </span> | ||
</div></html> | </div></html> | ||
| - | Among bacteria, Flagellin is a protein with highly conserved domains such as D0 and D1, whereas the sections D2 and D3 are extremely variable | + | Among bacteria, Flagellin is a protein with highly conserved domains such as D0 and D1, whereas the sections D2 and D3 are extremely variable (Yonekura et al. 2003). |
| - | Flagellin it is | + | By comparison of <i>Salmonella</i>, <i>Sphingomonas</i> and <i>Bacillus</i> |
| - | (Figure 2 | + | Flagellin it is remarkable that <i>Bacillus</i> D2 and D3 is reduced to a small loop |
| + | (Figure 2; Altegoer et al. 2014). | ||
<html> | <html> | ||
| - | <div class="figure" style="width: | + | <div class="figure" style="width:40%;"> |
<img src="https://static.igem.org/mediawiki/2014/1/12/MR_flagellen.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/1/12/MR_flagellen.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 2: Conserved protein domains of Flagellin among different bacteria. | + | <b>Figure 2: Conserved protein domains of Flagellin among different bacteria.</b> |
</span> | </span> | ||
</div></html> | </div></html> | ||
| - | This fact was the reason we took a closer look at the Flagellin of <i>Bacillus subtilis</i>. | + | This fact was the reason why we took a closer look at the Flagellin of <i>Bacillus subtilis</i>. |
| - | Our vision is | + | Our vision is to use the bacterial flagellum as a scaffold for high density accumulation of proteins with valuable functions. |
| - | accumulation of proteins with valuable functions. | + | |
“SURFing (Synthetic Units for Redirecting Functionalities) is the redirection of | “SURFing (Synthetic Units for Redirecting Functionalities) is the redirection of | ||
| Line 51: | Line 53: | ||
<img src="https://static.igem.org/mediawiki/2014/f/f9/MR_project_2.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/f/f9/MR_project_2.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 3: SURFing application workflow. | + | <b>Figure 3: SURFing application workflow.</b> |
</span> | </span> | ||
</div></html> | </div></html> | ||
| Line 59: | Line 61: | ||
Thus the SilverSURF project was born. | Thus the SilverSURF project was born. | ||
| - | <html><h2><a | + | <html><h2><a name="silver">SilverSURF</a></h2></html> |
| - | The purpose of the SilverSURF project is to catch ions from soil or solutions and | + | The purpose of the SilverSURF project is to catch ions from soil or solutions and sequester |
them with a high affinity. How should this be realized? Our vision is to introduce an | them with a high affinity. How should this be realized? Our vision is to introduce an | ||
additional modified copy of Flagellin into the genome of <i>Bacillus subtilis</i>. This allele | additional modified copy of Flagellin into the genome of <i>Bacillus subtilis</i>. This allele | ||
| Line 68: | Line 70: | ||
Flagellin is a chimeric protein with a new domain – the metal binding part of the metallothionein Cup1 from | Flagellin is a chimeric protein with a new domain – the metal binding part of the metallothionein Cup1 from | ||
yeast which specifically binds silver and copper ions (Moore et al. 2005). This leads | yeast which specifically binds silver and copper ions (Moore et al. 2005). This leads | ||
| - | to engineered <i>B. subtilis</i> cells, | + | to engineered <i>B. subtilis</i> cells, that catch the surrounding ions from their environment (Figure 4). |
<html> | <html> | ||
| Line 75: | Line 77: | ||
<img src="https://static.igem.org/mediawiki/2014/4/4e/MR_silver_1.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/4/4e/MR_silver_1.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 4: SilverSURF vision. | + | <b>Figure 4: SilverSURF vision.</b> |
</span> | </span> | ||
</div></html> | </div></html> | ||
| - | + | Many published data about transcriptional changes due to heavy metal ion stress in | |
| - | metal ion stress in <i>B. subtilis</i>; we were able to find specific promoters, which are more or less specific | + | <i>B. subtilis</i>; we were able to find specific promoters, which are more or less specific f |
| + | or certain metal ions. To analyze different promoters we | ||
established a reporter system. Therefore we used the promoter of | established a reporter system. Therefore we used the promoter of | ||
| - | <i>argJ</i> or the <i>arg</i>-operon respectively for sensing silver ions and the promoter of <i>ykuO</i> or its operon for the detection of copper ions, by generating a highly | + | <i>argJ</i> or the <i>arg</i>-operon respectively for sensing silver ions and the promoter of |
| + | <i>ykuO</i> or its operon for the detection of copper ions, by generating a highly | ||
modular <i>amyE</i> locus integration plasmid with a GFP reporter. By analyzing our reporter | modular <i>amyE</i> locus integration plasmid with a GFP reporter. By analyzing our reporter | ||
| - | constructs via STED microscopy, we | + | constructs via STED microscopy,we discovered that the chosen silver sensitive promoter does |
| - | does not show any differences by exposure to different silver concentrations compared | + | not show any differences by exposure to different silver concentrations compared to our negative |
| - | to our negative control without silver ions (Figure | + | control without silver ions (Figure 5). However, in principle the established |
promoter reporter system works though. | promoter reporter system works though. | ||
<html> | <html> | ||
| - | <div class="figure" style="width: | + | <div class="figure" style="width:70%;"> |
<img src="https://static.igem.org/mediawiki/2014/8/8b/Mr_project_silver_1.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/8/8b/Mr_project_silver_1.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 5: SilverSURF promoter reporter system with silver concentrations 0, 0.1 µM and 10 µM. | + | <b>Figure 5: SilverSURF promoter reporter system with silver concentrations 0, 0.1 µM and 10 µM.</b> |
</span> | </span> | ||
</div></html> | </div></html> | ||
| - | Nevertheless we focused | + | Nevertheless we focused on the Flagellin Cup1 chimera. We designed |
a modular system for chromosomal integration, where the <i>hag</i> gene, which codes for Flagellin, | a modular system for chromosomal integration, where the <i>hag</i> gene, which codes for Flagellin, | ||
features a <i>Spe</i>I site for fast and modular insertion of domains via Gibson Assembly (pIGEM-0016). | features a <i>Spe</i>I site for fast and modular insertion of domains via Gibson Assembly (pIGEM-0016). | ||
| - | The Cup1 module was amplified from <i>Saccharomyces cerevisiae</i> genomic DNA integrated into the | + | The Cup1 module was amplified from <i>Saccharomyces cerevisiae</i> genomic DNA and integrated into the pIGEM-0016 system. In the end the wild type |
| - | pIGEM-0016 system | + | <i>hag</i> was successfully replaced with the chimeric <i>hag-cup1</i> |
| - | construct. pMAD allows high efficiency allelic replacement of the wild type <i>hag</i> via the | + | construct <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">(BBa_K1329002)</a></html>. |
| + | pMAD allows high efficiency allelic replacement of the wild type <i>hag</i> via the | ||
pMAD system (Arnaud et al. 2004). | pMAD system (Arnaud et al. 2004). | ||
| - | <html><h2><a | + | <html><h2><a name="motility">Motility assays and structural analysis</a></h2></html> |
As an <i>in vivo</i> functional test of the engineered Flagellin, swimming and swarming | As an <i>in vivo</i> functional test of the engineered Flagellin, swimming and swarming | ||
assays were performed. Unfortunately the assays showed that the engineered Flagellin | assays were performed. Unfortunately the assays showed that the engineered Flagellin | ||
containing strains were strongly reduced in their motility compared to both the wild | containing strains were strongly reduced in their motility compared to both the wild | ||
| - | type strain WT3610 and the control strain producing our modularized Flagellin Hag-<i>Spe</i>I | + | type strain WT3610 and the control strain producing our modularized Flagellin Hag-<i>Spe</i>I <html><a href="" target="_blank">(BBa_K1329003)</a></html> |
without the inserted <i>cup1</i> domain. | without the inserted <i>cup1</i> domain. | ||
| - | Anyway, to | + | Anyway, to learned what might disrupt the flagellum functionality. The <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank"><i>hag-cup1</i></a></html> was |
| - | overexpressed in <i>Escherichia coli</i> | + | overexpressed in <i>Escherichia coli</i>and sufficient amounts of protein could be purified, |
which were subjected to crystallization to solve the structure of the chimeric protein. | which were subjected to crystallization to solve the structure of the chimeric protein. | ||
Crystals could be obtained but the structure could only be solved partially, due to the disruption | Crystals could be obtained but the structure could only be solved partially, due to the disruption | ||
| Line 123: | Line 128: | ||
These results led us to rethink our approach and we concluded that the Flagellin might be functional if | These results led us to rethink our approach and we concluded that the Flagellin might be functional if | ||
we replace the <i>Bacillus</i> loop with the D2 Domain of <i>Salmonella typhimurium</i> (Figure 2) and | we replace the <i>Bacillus</i> loop with the D2 Domain of <i>Salmonella typhimurium</i> (Figure 2) and | ||
| - | use it as a linker for the Cup1 (Figure 6). This construct is further on called Hag-D2-Cup1. | + | use it as a linker for the Cup1 (Figure 6). This construct is further on called |
| + | <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a></html>. | ||
<html> | <html> | ||
| - | <div class="figure" style="width: | + | <div class="figure" style="width:1%;"> |
<img src="https://static.igem.org/mediawiki/2014/e/e0/MR_cup_flagellin.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/e/e0/MR_cup_flagellin.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 6: New engineered synthetic Hag-D2-Cup1 Flagellin. | + | <b>Figure 6: New engineered synthetic <a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a> Flagellin.</b> |
</span> | </span> | ||
</div></html> | </div></html> | ||
| Line 138: | Line 144: | ||
further information see the <html><a href="https://2014.igem.org/Team:Marburg:Project:Tumor">CancerSURF</a></html> project). | further information see the <html><a href="https://2014.igem.org/Team:Marburg:Project:Tumor">CancerSURF</a></html> project). | ||
| - | The Hag-D2-Cup1 and Hag-D2-Strep were also integrated into <i>B. subtilis</i> via homologous | + | The <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a></html> and Hag-D2-Strep |
| + | <html><a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">(BBa_K1329003)</a></html> | ||
| + | were also integrated into <i>B. subtilis</i> via homologous | ||
recombination by using our modular pMAD system. Again motility assays were | recombination by using our modular pMAD system. Again motility assays were | ||
| - | performed, which showed comparable results for Hag-Cup1 and Hag-D2-Cup1 though the strain containing Hag-D2-Strep behaves similar to the wild type in this assay (Figure 7). | + | performed, which showed comparable results for <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-Cup1</a></html> |
| + | and <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a></html> though the strain containing | ||
| + | <html><a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">Hag-D2-Strep</a></html> behaves similar to the wild type in this assay (Figure 7). | ||
<html> | <html> | ||
| Line 146: | Line 156: | ||
<img src="https://static.igem.org/mediawiki/2014/0/09/Mr_project_silver_2.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/0/09/Mr_project_silver_2.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 7: Motility assays of the Hag-D2-Cup1 and the Hag-D2-Strep. | + | <b>Figure 7: Motility assays of the <a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a> and the |
| + | <a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">Hag-D2-Strep</a> mutants.</b> | ||
</span> | </span> | ||
</div></html> | </div></html> | ||
| - | We asked ourselves how | + | We asked ourselves how one construct can show a wild type like phenotype whereas the other doesn't. |
| - | phenotype | + | Therefore, we visualized the flagella by using electron microscopy |
with an uranyl acetate negative staining of our different <i>Bacillus</i> strains (Figure 8). To our surprise | with an uranyl acetate negative staining of our different <i>Bacillus</i> strains (Figure 8). To our surprise | ||
| - | we could show that the Hag-D2-Cup1 strain is able to form filaments though they | + | we could show that the <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a></html> strain is able to form filaments though they |
are much shorter and inflexible, compared to the wild type flagella, which might | are much shorter and inflexible, compared to the wild type flagella, which might | ||
| - | be the reason why we observed fragmented filaments. Astonishingly the Hag-D2-Strep | + | be the reason why we observed fragmented filaments. Astonishingly the <html><a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">Hag-D2-Strep</a></html> |
| - | filaments looked like wild type filaments. Although we observed less filaments they | + | filaments looked like wild type filaments. Although we observed less filaments they seemed to be functional. |
| - | to be functional. What might be the difference between Hag-D2-Cup1 and Hag-D2-Strep | + | What might be the difference between <html><a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a></html> and |
| + | <html><a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">Hag-D2-Strep</a></html> | ||
is part of further investigations. A major factor could be the smaller size of the StrepTag, which consists of only eight amino acids, in contrast to the 4.5-fold bigger part of the Cup1 (36 amino acids). | is part of further investigations. A major factor could be the smaller size of the StrepTag, which consists of only eight amino acids, in contrast to the 4.5-fold bigger part of the Cup1 (36 amino acids). | ||
| Line 164: | Line 176: | ||
<img src="https://static.igem.org/mediawiki/2014/2/26/Mr_team_EMSURF.png" alt="" /> | <img src="https://static.igem.org/mediawiki/2014/2/26/Mr_team_EMSURF.png" alt="" /> | ||
<span class="caption"> | <span class="caption"> | ||
| - | Figure 8: Uranylacetate negative stained electron microscopy of <i>B. subtilis</i> (A) and | + | <b>Figure 8: Uranylacetate negative stained electron microscopy of <i>B. subtilis</i> (A) and |
| + | <a href="http://parts.igem.org/Part:BBa_K1329002" target="_blank">Hag-D2-Cup1</a> strain (B).</b> | ||
</span> | </span> | ||
</div></html> | </div></html> | ||
| - | The successful engineering of the Hag-D2-Strep Flagellin is the cornerstone for the success story of our | + | The successful engineering of the <html><a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">Hag-D2-Strep</a></html> |
| - | CancerSURF project. In further investigations we plan to couple Cup1-Streptavidin, which was successfully produced recombinantly, to isolated Hag-D2-Strep filaments | + | Flagellin is the cornerstone for the success story of our |
| + | CancerSURF project. In further investigations we plan to couple Cup1-Streptavidin, which was successfully produced recombinantly, to isolated | ||
| + | <html><a href="http://parts.igem.org/Part:BBa_K1329003" target="_blank">Hag-D2-Strep</a></html> filaments | ||
for our SilverSURF – this would directly optimize our approach for pollution reduction. Beyond that, | for our SilverSURF – this would directly optimize our approach for pollution reduction. Beyond that, | ||
there is no need to use the GMO outside the laboratory. The isolated recombinant flagella could be used directly so no major safety concerns would emerge. Also the higher level of modularity allows us to couple every | there is no need to use the GMO outside the laboratory. The isolated recombinant flagella could be used directly so no major safety concerns would emerge. Also the higher level of modularity allows us to couple every | ||
imaginable high value applicable protein to our filaments. There is no reason to use only one protein, we | imaginable high value applicable protein to our filaments. There is no reason to use only one protein, we | ||
also plan to use different proteins coupled to streptavidin and bring them into close proximity by using | also plan to use different proteins coupled to streptavidin and bring them into close proximity by using | ||
| - | the filaments as a scaffold for molecular crowding. There | + | the filaments as a scaffold for molecular crowding. There is a huge repertoire of valuable applications for |
| - | this system to solve all kinds of challenges | + | this system to solve all kinds of challenges humanity has to face. |
<html><hr /></html> | <html><hr /></html> | ||
Latest revision as of 02:22, 18 October 2014
SilverSURF
Introduction
"Surfing is a surface water sport in which the wave rider, referred to as a surfer, rides on the forward or deep face of a moving wave, which is usually carrying the surfer towards the shore."
In modern science, at the limits of visualization and detection feasibility, the
amplification of signals is a serious challenge. Scaffolding – building a high
density of factors in close proximity – is the key to optimize chemical reactions or
increasing signals for detection. Here we use the bacterial flagellum, a structure provided by nature itself,
as a toolbox for different applications. This huge formation consists
of the basal body, the hook and the filament, which is composed of up to 20.000 copies of the protein Flagellin (Figure 1). Due to the rotating flagellum, bacteria are able to
"surf" through their environment.
 Figure 1: Composition of the bacterial flagellar filament.
Figure 1: Composition of the bacterial flagellar filament.
Among bacteria, Flagellin is a protein with highly conserved domains such as D0 and D1, whereas the sections D2 and D3 are extremely variable (Yonekura et al. 2003). By comparison of Salmonella, Sphingomonas and Bacillus Flagellin it is remarkable that Bacillus D2 and D3 is reduced to a small loop (Figure 2; Altegoer et al. 2014).
 Figure 2: Conserved protein domains of Flagellin among different bacteria.
Figure 2: Conserved protein domains of Flagellin among different bacteria.
This fact was the reason why we took a closer look at the Flagellin of Bacillus subtilis. Our vision is to use the bacterial flagellum as a scaffold for high density accumulation of proteins with valuable functions.
“SURFing (Synthetic Units for Redirecting Functionalities) is the redirection of natural occurring structures to a high density surface which can be modified for different purposes, towards solving important challenges for humanity e.g. reduction of environmental pollution and improvement or development of new medical applications (Figure 3).”
 Figure 3: SURFing application workflow.
Figure 3: SURFing application workflow.
Utilizing the principle of SURF on the basis of bacterial flagella, we decided to contribute to the reduction of environmental pollution, concerning heavy metal ions, some of which expose a high toxicity and are harmful even in small doses. A well known representative of these kind of metals is silver (Ag). The toxic effect is on the one hand industrially welcome, as silver is used as an additive to kill bacteria on the other hand it is a big problem due to its dissemination by industrial waste water. Thus the SilverSURF project was born.
SilverSURF
The purpose of the SilverSURF project is to catch ions from soil or solutions and sequester them with a high affinity. How should this be realized? Our vision is to introduce an additional modified copy of Flagellin into the genome of Bacillus subtilis. This allele should be controlled by a silver ion sensitive promoter. In presence of silver ions the modified Flagellin is expressed and incorporated into the filament. This modified Flagellin is a chimeric protein with a new domain – the metal binding part of the metallothionein Cup1 from yeast which specifically binds silver and copper ions (Moore et al. 2005). This leads to engineered B. subtilis cells, that catch the surrounding ions from their environment (Figure 4).

 Figure 4: SilverSURF vision.
Figure 4: SilverSURF vision.
Many published data about transcriptional changes due to heavy metal ion stress in B. subtilis; we were able to find specific promoters, which are more or less specific f or certain metal ions. To analyze different promoters we established a reporter system. Therefore we used the promoter of argJ or the arg-operon respectively for sensing silver ions and the promoter of ykuO or its operon for the detection of copper ions, by generating a highly modular amyE locus integration plasmid with a GFP reporter. By analyzing our reporter constructs via STED microscopy,we discovered that the chosen silver sensitive promoter does not show any differences by exposure to different silver concentrations compared to our negative control without silver ions (Figure 5). However, in principle the established promoter reporter system works though.
 Figure 5: SilverSURF promoter reporter system with silver concentrations 0, 0.1 µM and 10 µM.
Figure 5: SilverSURF promoter reporter system with silver concentrations 0, 0.1 µM and 10 µM.
Nevertheless we focused on the Flagellin Cup1 chimera. We designed a modular system for chromosomal integration, where the hag gene, which codes for Flagellin, features a SpeI site for fast and modular insertion of domains via Gibson Assembly (pIGEM-0016). The Cup1 module was amplified from Saccharomyces cerevisiae genomic DNA and integrated into the pIGEM-0016 system. In the end the wild type hag was successfully replaced with the chimeric hag-cup1 construct (BBa_K1329002). pMAD allows high efficiency allelic replacement of the wild type hag via the pMAD system (Arnaud et al. 2004).
Motility assays and structural analysis
As an in vivo functional test of the engineered Flagellin, swimming and swarming assays were performed. Unfortunately the assays showed that the engineered Flagellin containing strains were strongly reduced in their motility compared to both the wild type strain WT3610 and the control strain producing our modularized Flagellin Hag-SpeI (BBa_K1329003) without the inserted cup1 domain.
Anyway, to learned what might disrupt the flagellum functionality. The hag-cup1 was overexpressed in Escherichia coliand sufficient amounts of protein could be purified, which were subjected to crystallization to solve the structure of the chimeric protein. Crystals could be obtained but the structure could only be solved partially, due to the disruption of the D1 domain by the cup1 integration into hag.
These results led us to rethink our approach and we concluded that the Flagellin might be functional if we replace the Bacillus loop with the D2 Domain of Salmonella typhimurium (Figure 2) and use it as a linker for the Cup1 (Figure 6). This construct is further on called Hag-D2-Cup1.
 Figure 6: New engineered synthetic Hag-D2-Cup1 Flagellin.
Figure 6: New engineered synthetic Hag-D2-Cup1 Flagellin.
At the same time we developed the idea to insert a StrepTag into the Hag-D2 construct which would provide an enormous flexibility by coupling a multitude of functional proteins to streptavidin and further on build extracellular complexes of Flagellin with chimeric streptavidin proteins (for further information see the CancerSURF project).
The Hag-D2-Cup1 and Hag-D2-Strep (BBa_K1329003) were also integrated into B. subtilis via homologous recombination by using our modular pMAD system. Again motility assays were performed, which showed comparable results for Hag-Cup1 and Hag-D2-Cup1 though the strain containing Hag-D2-Strep behaves similar to the wild type in this assay (Figure 7).
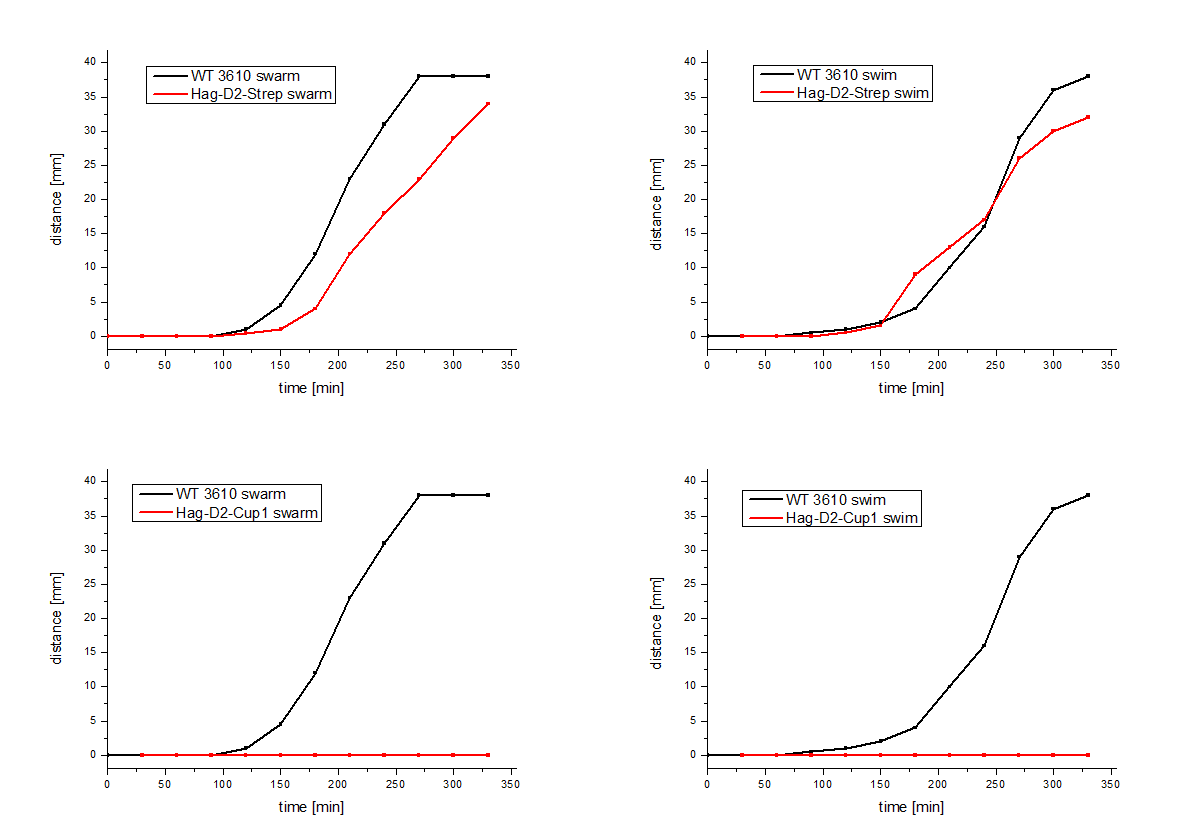 Figure 7: Motility assays of the Hag-D2-Cup1 and the
Hag-D2-Strep mutants.
Figure 7: Motility assays of the Hag-D2-Cup1 and the
Hag-D2-Strep mutants.
We asked ourselves how one construct can show a wild type like phenotype whereas the other doesn't. Therefore, we visualized the flagella by using electron microscopy with an uranyl acetate negative staining of our different Bacillus strains (Figure 8). To our surprise we could show that the Hag-D2-Cup1 strain is able to form filaments though they are much shorter and inflexible, compared to the wild type flagella, which might be the reason why we observed fragmented filaments. Astonishingly the Hag-D2-Strep filaments looked like wild type filaments. Although we observed less filaments they seemed to be functional. What might be the difference between Hag-D2-Cup1 and Hag-D2-Strep is part of further investigations. A major factor could be the smaller size of the StrepTag, which consists of only eight amino acids, in contrast to the 4.5-fold bigger part of the Cup1 (36 amino acids).
 Figure 8: Uranylacetate negative stained electron microscopy of B. subtilis (A) and
Hag-D2-Cup1 strain (B).
Figure 8: Uranylacetate negative stained electron microscopy of B. subtilis (A) and
Hag-D2-Cup1 strain (B).
The successful engineering of the Hag-D2-Strep Flagellin is the cornerstone for the success story of our CancerSURF project. In further investigations we plan to couple Cup1-Streptavidin, which was successfully produced recombinantly, to isolated Hag-D2-Strep filaments for our SilverSURF – this would directly optimize our approach for pollution reduction. Beyond that, there is no need to use the GMO outside the laboratory. The isolated recombinant flagella could be used directly so no major safety concerns would emerge. Also the higher level of modularity allows us to couple every imaginable high value applicable protein to our filaments. There is no reason to use only one protein, we also plan to use different proteins coupled to streptavidin and bring them into close proximity by using the filaments as a scaffold for molecular crowding. There is a huge repertoire of valuable applications for this system to solve all kinds of challenges humanity has to face.
Altegoer, Florian; Schuhmacher, Jan; Pausch, Patrick; Bange, Gert (2014): From molecular evolution to biobricks and synthetic modules: a lesson by the bacterial flagellum. In: Biotechnology and Genetic Engineering Reviews 30 (1), S. 49–64. DOI: 10.1080/02648725.2014.921500.
Arnaud, Maryvonne; Chastanet, Arnaud; Débarbouillé, Michel (2004): New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. In: Appl. Environ. Microbiol. 70 (11), S. 6887–6891. DOI: 10.1128/AEM.70.11.6887-6891.2004.
Moore, Charles M.; Gaballa, Ahmed; Hui, Monica; Ye, Rick W.; Helmann, John D. (2005): Genetic and physiological responses of Bacillus subtilis to metal ion stress. In: Molecular Microbiology 57 (1), S. 27–40, zuletzt geprüft am 16.10.2014.
Yonekura, Koji; Maki-Yonekura, Saori; Namba, Keiichi (2003): Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. In: Nature 424 (6949), S. 643–650. DOI: 10.1038/nature01830.
 "
"