Team:ETH Zurich/expresults/integrases
From 2014.igem.org
(→Integrases) |
(→Integrases) |
||
| (31 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
| - | In order to characterize the integrase system described above, we first combined the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] with one of our integrase genes | + | In order to characterize the integrase system described above, we first combined the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] with one of our [https://2014.igem.org/Team:ETH_Zurich/modeling/int integrase] genes ''bxb1'', followed directly by a red fluorescent protein (RFP, mCherry) to make the expression accessible. Also, this system includes an XOR buffer gate per default blocking transcription of sfGFP. Upon BXB1 activity and switching the gate into ON-state, the terminator should have been removed and sfGFP should have been expressed. We intially designed three different constructs for characterization of the recombinases and their cross-activity. However, the measurement of fluorescent proteins, with both a [https://2014.igem.org/Team:ETH_Zurich/lab/protocols#Tecan_Infinite_M200_Pro.E2.84.A2 plate reader] and a [https://2014.igem.org/Team:ETH_Zurich/lab/protocols#BD_LSRFortessa.E2.84.A2_Flow_Cytometer_System flow cytometer], did not indicate sfGFP expression due to recombinase activity. Nevertheless, RFP was clearly detectable upon induction in plate reader experiments suggesting that the induction itself worked (see figure 5 and 6). |
{|class="wikitable resized" style="background-color: white; border: 0px+important;" | {|class="wikitable resized" style="background-color: white; border: 0px+important;" | ||
| - | |[[File:ETH Zurich 2014 250min Integrase mCherry.png|center|600px|thumb|''' | + | |[[File:ETH Zurich 2014 250min Integrase mCherry corrected.png|center|600px|thumb|'''Figure 5''' '''Expression of red fluorescent protein (RFP) and green fluorescent protein (GFP) 250 min after induction with various concentrations of 3OC6-HSL (10<sup>-13</sup> M to 10<sup>-4</sup> M).''' RFP is under the control of the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] together with the integrase BXB1. Upon expression of BXB1 a buffer gate should have been swtiched to ON-state producing GFP. Data points are mean values of triplicate measurements in 96-well microtiter plates ± standard deviation. For the full data set and kinetics please [https://2014.igem.org/Team:ETH_Zurich/contact contact] us or visit the [https://2014.igem.org/Team:ETH_Zurich/data/raw raw data] page.]] |
| - | |[[File:ETH Zurich 2014 610minIntegrase.png|center|600px|thumb|''' | + | |[[File:ETH Zurich 2014 610minIntegrase corrected.png|center|600px|thumb|'''Figure 6''' '''Expression of red fluorescent protein (RFP) and green fluorescent protein (GFP) 610 min after induction with various concentrations of 3OC6-HSL (10<sup>-13</sup> M to 10<sup>-4</sup> M).''' RFP is under the control of the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] together with the integrase BXB1. Upon expression of BXB1 a buffer gate should have been swtiched to ON-state producing GFP. Data points are mean values of triplicate measurements in 96-well microtiter plates ± standard deviation. For the full data set and kinetics please [https://2014.igem.org/Team:ETH_Zurich/contact contact] us or visit the [https://2014.igem.org/Team:ETH_Zurich/data/raw raw data] page.]] |
|} | |} | ||
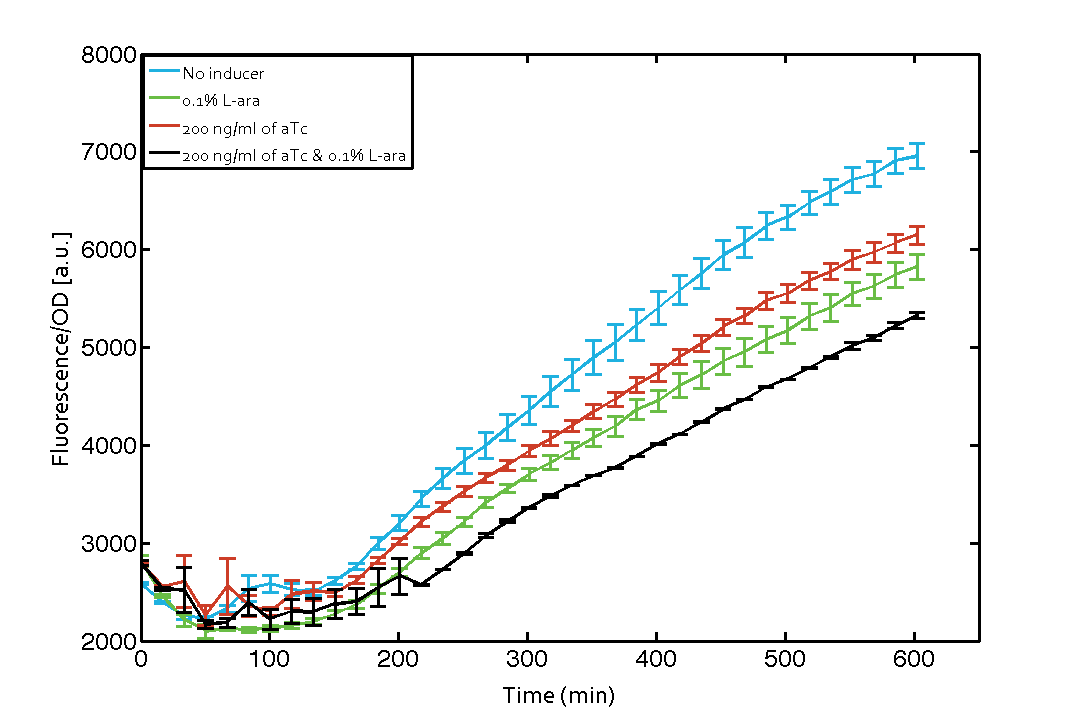
| - | As our constructs did not show the expected functionality, we decided to directly use the plasmids described by Bonnet ''et al''.<sup>[[Team:ETH_Zurich/project/references#refBonnet|[9]]]</sup> which where obtained from addgene ([http://www.addgene.org/44456 Dual-recombinase-controller], [http://www.addgene.org/44453/ XOR gate-V2.0]). However, we were using a TOP10 strain not expressing TetR by default (as compared to DH5alphaZ1) and as a result our strain had to be co-transformed with an additional plasmid encoding TetR. Also, we used defined [https://2014.igem.org/Team:ETH_Zurich/lab/protocols#Defined_M9_medium_with_0.4_.25_glycerol M9 medium] with 0.4% glycerol and 1% CAA instead of proprietary defined medium ([http://teknova-blog.com/hi-def-azure-media Teknova Hi-Def Azure medium]). As of today, this set-up did not allow us to get the integrase XOR gate running. We are not giving up on this and are proceeding with debugging our construct further and hope to find a solution until the Giant Jamboree in Boston. | + | As our constructs did not show the expected functionality, we decided to directly use the plasmids described by Bonnet ''et al''.<sup>[[Team:ETH_Zurich/project/references#refBonnet|[9]]]</sup> which where obtained from addgene ([http://www.addgene.org/44456 Dual-recombinase-controller], [http://www.addgene.org/44453/ XOR gate-V2.0]). The data available from the original experiment by Bonnet<sup>[[Team:ETH_Zurich/project/references#refBonnet|[9]]]</sup> was used in our model to retrieve the [https://2014.igem.org/Team:ETH_Zurich/modeling/int missing parameters of integrases]. However, we were using a [http://www.openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] strain not expressing TetR by default (as compared to [http://www.expressys.com/main_strains.html DH5alphaZ1]) and as a result our strain had to be co-transformed with an additional plasmid encoding TetR. Also, we used defined [https://2014.igem.org/Team:ETH_Zurich/lab/protocols#Defined_M9_medium_with_0.4_.25_glycerol M9 medium] with 0.4% glycerol and 1% CAA instead of proprietary defined medium ([http://teknova-blog.com/hi-def-azure-media Teknova Hi-Def Azure medium]). As of today, this set-up did not allow us to get the integrase XOR gate running. The fluorescence readout for ON-states (exactly one input, either 0.1% L-arabinose or 200 ng/mL anhydrous tetracycline) and the OFF-states was not as expected (see figure 7). The OFF-state should not show increased fluorescence over time, while the ON-states should increase significantly after 4 h and continue to increase over the whole time span<sup>[[Team:ETH_Zurich/project/references#refBonnet|[9]]]</sup>. We are not giving up on this and are proceeding with debugging our construct further and hope to find a solution until the Giant Jamboree in Boston. |
| + | |||
| + | |||
| + | {{:Team:ETH_Zurich/tpl/topbutton|green}} | ||
| + | |||
| + | |||
| + | [[File:ETH Zurich 2014 integrases addgene plasmids corrected.png|center|600px|thumb|'''Figure 7 Expression of green fluorescent protein (GFP) after induction of [https://2014.igem.org/Team:ETH_Zurich/modeling/xor#Biological_Principles XOR integrase logic].''' Upon induction of integrase BXB1 (t=0 min) a [https://2014.igem.org/Team:ETH_Zurich/lab/sequences#BUFFER_Gate_Construct buffer gate] should have been switched to ON-state producing GFP only if either L-arabinose (L-ara) or anhydrous tetracycline (aTc) is present. Data points are mean values of triplicate measurements in 96-well microtiter plates ± standard deviation. For the full data set and kinetics please [https://2014.igem.org/Team:ETH_Zurich/contact contact] us or visit the [https://2014.igem.org/Team:ETH_Zurich/data/raw raw data] page.]] | ||
Latest revision as of 01:19, 18 October 2014
Integrases
The design of our XOR gates was based on integrase logic[9]. This means, depending on the input molecules, integrases can be expressed, subsequently switch a terminator sequence previously blocking gene expression, and then the output gene can be transcribed. This approach is explained here.
In order to characterize the integrase system described above, we first combined the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] with one of our integrase genes bxb1, followed directly by a red fluorescent protein (RFP, mCherry) to make the expression accessible. Also, this system includes an XOR buffer gate per default blocking transcription of sfGFP. Upon BXB1 activity and switching the gate into ON-state, the terminator should have been removed and sfGFP should have been expressed. We intially designed three different constructs for characterization of the recombinases and their cross-activity. However, the measurement of fluorescent proteins, with both a plate reader and a flow cytometer, did not indicate sfGFP expression due to recombinase activity. Nevertheless, RFP was clearly detectable upon induction in plate reader experiments suggesting that the induction itself worked (see figure 5 and 6).
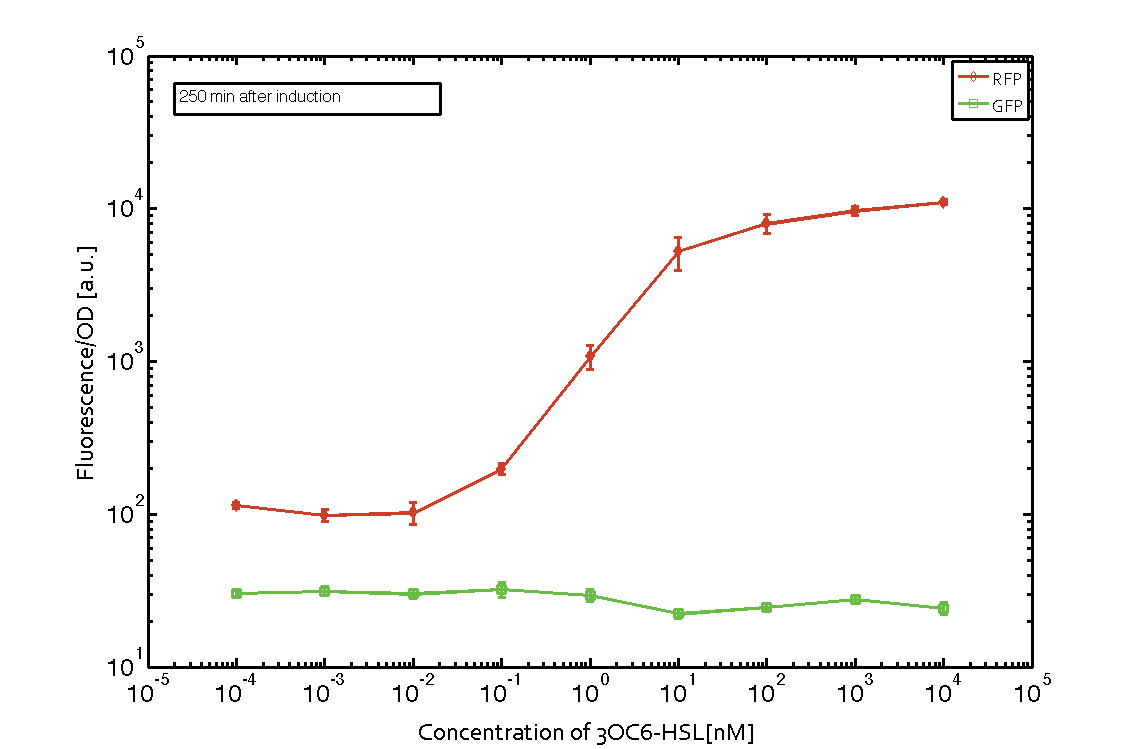 Figure 5 Expression of red fluorescent protein (RFP) and green fluorescent protein (GFP) 250 min after induction with various concentrations of 3OC6-HSL (10-13 M to 10-4 M). RFP is under the control of the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] together with the integrase BXB1. Upon expression of BXB1 a buffer gate should have been swtiched to ON-state producing GFP. Data points are mean values of triplicate measurements in 96-well microtiter plates ± standard deviation. For the full data set and kinetics please contact us or visit the raw data page. | 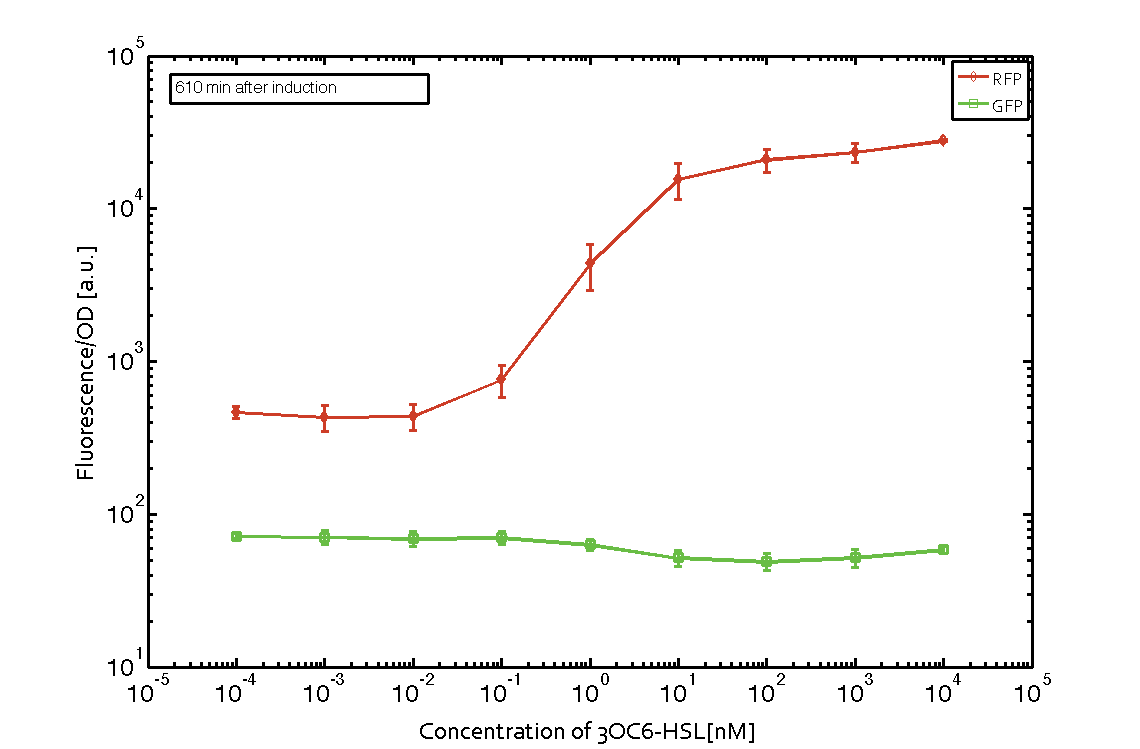 Figure 6 Expression of red fluorescent protein (RFP) and green fluorescent protein (GFP) 610 min after induction with various concentrations of 3OC6-HSL (10-13 M to 10-4 M). RFP is under the control of the [http://parts.igem.org/Part:BBa_R0062 pLux promoter (BBa_R0062)] together with the integrase BXB1. Upon expression of BXB1 a buffer gate should have been swtiched to ON-state producing GFP. Data points are mean values of triplicate measurements in 96-well microtiter plates ± standard deviation. For the full data set and kinetics please contact us or visit the raw data page. |
As our constructs did not show the expected functionality, we decided to directly use the plasmids described by Bonnet et al.[9] which where obtained from addgene ([http://www.addgene.org/44456 Dual-recombinase-controller], [http://www.addgene.org/44453/ XOR gate-V2.0]). The data available from the original experiment by Bonnet[9] was used in our model to retrieve the missing parameters of integrases. However, we were using a [http://www.openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] strain not expressing TetR by default (as compared to [http://www.expressys.com/main_strains.html DH5alphaZ1]) and as a result our strain had to be co-transformed with an additional plasmid encoding TetR. Also, we used defined M9 medium with 0.4% glycerol and 1% CAA instead of proprietary defined medium ([http://teknova-blog.com/hi-def-azure-media Teknova Hi-Def Azure medium]). As of today, this set-up did not allow us to get the integrase XOR gate running. The fluorescence readout for ON-states (exactly one input, either 0.1% L-arabinose or 200 ng/mL anhydrous tetracycline) and the OFF-states was not as expected (see figure 7). The OFF-state should not show increased fluorescence over time, while the ON-states should increase significantly after 4 h and continue to increase over the whole time span[9]. We are not giving up on this and are proceeding with debugging our construct further and hope to find a solution until the Giant Jamboree in Boston.
 "
"