Team:Oxford/biosensor characterisation
From 2014.igem.org
(Difference between revisions)
Olivervince (Talk | contribs) |
|||
| (138 intermediate revisions not shown) | |||
| Line 29: | Line 29: | ||
</div> | </div> | ||
<div style="background-color:white; border-bottom-left-radius:10px;border-radius:10px; padding-left:10px;padding-right:10px;min-width:300px;margin-top:-50px;"> | <div style="background-color:white; border-bottom-left-radius:10px;border-radius:10px; padding-left:10px;padding-right:10px;min-width:300px;margin-top:-50px;"> | ||
| - | <a href=" | + | <a href="https://static.igem.org/mediawiki/2014/3/3d/OxigemLAB_BOOK.pdf" target="_blank"><img src="https://static.igem.org/mediawiki/2014/5/50/OxigemLabbook.png" style="position:absolute;width:6%;margin-left:84%;margin-top:-13%;z-index:10;"></a> |
<a href="https://static.igem.org/mediawiki/2014/1/16/Oxigem_LAB_PROTOCOLS.pdf" target="_blank"><img src="https://static.igem.org/mediawiki/2014/a/a4/OxigemProtocols.png" style="position:absolute;width:6%;margin-left:91%;margin-top:-13%;z-index:10;"></a> | <a href="https://static.igem.org/mediawiki/2014/1/16/Oxigem_LAB_PROTOCOLS.pdf" target="_blank"><img src="https://static.igem.org/mediawiki/2014/a/a4/OxigemProtocols.png" style="position:absolute;width:6%;margin-left:91%;margin-top:-13%;z-index:10;"></a> | ||
| Line 36: | Line 36: | ||
<div style="width:100%;"><font style="font-size:15px;font-weight:500;">Show all:</font></div> | <div style="width:100%;"><font style="font-size:15px;font-weight:500;">Show all:</font></div> | ||
| - | <a href="#showmodelling"><div class="orange_news_block1 showmodelling" style="background: #F9A7B0;border-radius:15px;color:black;float:left;height:40%;width:40%;margin-left:6%;padding-top: | + | <a href="#showmodelling"><div class="orange_news_block1 showmodelling" style="background: #F9A7B0;border-radius:15px;color:black;float:left;height:40%;width:40%;margin-left:6%;padding-top:10px;"><center> |
| - | <h1white><font style="font-size:15px;font-weight:500;">Modelling</font> | + | <h1white><font style="font-size:15px;font-weight:500;">Modelling</font></h1white></center> |
</div></a> | </div></a> | ||
| - | <a href="#showwetlab"><div class="orange_news_block1 showwetlab" style="background: #ADD8E6;border-radius:15px;color:black;float:left;height:40%;width:40 | + | <a href="#showwetlab"><div class="orange_news_block1 showwetlab" style="background: #ADD8E6;border-radius:15px;color:black;float:left;height:40%;width:40%;margin-left:3%;padding-top:10px;"><center> |
| - | <h1white><font style="font-size:15px;font-weight:500;">Wetlab</font> | + | <h1white><font style="font-size:15px;font-weight:500;">Wetlab</font></h1white></center> |
</div></a> | </div></a> | ||
<br><br><br><br><br> | <br><br><br><br><br> | ||
| Line 49: | Line 49: | ||
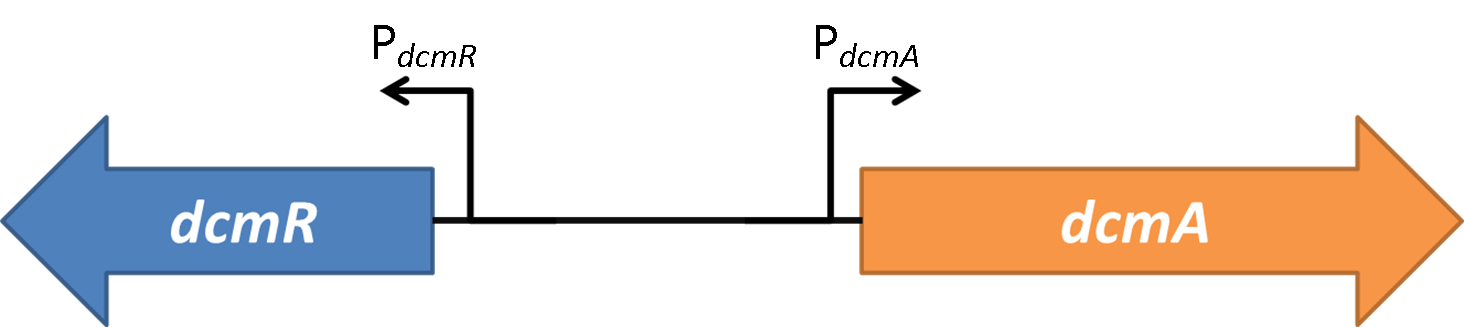
<font style="font-style: italic;">Methylobacterium Extorquens</font> DM4 in the presence of DCM expresses DcmA, a dichloromethane dehalogenase. | <font style="font-style: italic;">Methylobacterium Extorquens</font> DM4 in the presence of DCM expresses DcmA, a dichloromethane dehalogenase. | ||
| - | Within 1.5kb upstream of <font style="font-style: italic;">dcmA</font> and in the opposite orientation is a second gene encoding DcmR, a regulatory protein that controls expression of | + | Within 1.5kb upstream of <font style="font-style: italic;">dcmA</font> and in the opposite orientation is a second gene encoding DcmR, a regulatory protein that controls expression of <font style="font-style: italic;">dcmA</font>.<br><br> |
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/7/7f/Oxford_charac3.png" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> | ||
| + | <br><br> | ||
| + | |||
| - | |||
| - | In order to design and create a stable and sensitive system that responds to DCM we first need to characterise the regulatory nature of DcmR. Characterisation of this regulatory network has never been done before although it has been suggested to be a repressor [1]; we will be the first to fully characterise the mode of action of <font style="font-style: italic;">dcmR</font>. To do this we | + | In order to design and create a stable and sensitive system that responds to DCM we first need to characterise the regulatory nature of DcmR. Characterisation of this regulatory network has never been done before although it has been suggested to be a repressor [1]; we will be the first to fully characterise the mode of action of <font style="font-style: italic;">dcmR</font>. To do this we are testing the following hypotheses for DCM activating the transcription of <font style="font-style: italic;">dcmR</font>: either double repression or double activation. In other words, either DcmR represses <font style="font-style: italic;">dcmA</font> expression and DcmR is in negatively modulated by the presence of DCM; or expression of <font style="font-style: italic;">dcmA</font> requires DcmR as an activator, with DcmR in turn only activated in the presence of DCM.<br><br> |
</div> | </div> | ||
| Line 88: | Line 91: | ||
<h1>DcmR and regulation of <font style="font-style: italic;">dcmA</font> expression</h1> | <h1>DcmR and regulation of <font style="font-style: italic;">dcmA</font> expression</h1> | ||
| - | Mutants with <font style="font-style: italic;">dcmA</font> and the intergenic region but without complete <font style="font-style: italic;"> | + | Mutants with <font style="font-style: italic;">dcmA</font> and the intergenic region but without complete DcmR express <font style="font-style: italic;">dcmA</font> constitutively. Re-integration of <font style="font-style: italic;">dcmR</font> restores regulation of <font style="font-style: italic;">dcmA</font> expression at the transcriptional level [1]. In addition, it has been shown that the region including <font style="font-style: italic;">dcmR</font>, the intergenic region and <font style="font-style: italic;">dcmA</font> is sufficient to confer a DCM dependent response in genetically engineered <font style="font-style: italic;">Methylobacterium extorquens DM4</font> [2]. <br><br> |
<h1>DcmR and DNA-binding</h1> | <h1>DcmR and DNA-binding</h1> | ||
| - | DcmR is thought to be a DNA binding protein as structure predicting software indicates that there is a helix-turn-helix domain at the N-terminal of the protein. Since the region between the two promoters for <font style="font-style: italic;">dcmR</font> and <font style="font-style: italic;">dcmA</font> can be deleted without any effect on regulation it has been suggested that DcmR does not to a secondary regulatory site in between the genes but most likely acts directly on the <font style="font-style: italic;">dcmA</font> promoter itself [1]. In addition, regulated expression of <font style="font-style: italic;">dcmA</font> is not | + | DcmR is thought to be a DNA binding protein as structure predicting software indicates that there is a helix-turn-helix domain at the N-terminal of the protein. Since the region between the two promoters for <font style="font-style: italic;">dcmR</font> and <font style="font-style: italic;">dcmA</font> can be deleted without any effect on regulation it has been suggested that DcmR does not bind to a secondary regulatory site in between the genes but most likely acts directly on the <font style="font-style: italic;">dcmA</font> promoter itself [1]. In addition, regulated expression of <font style="font-style: italic;">dcmA</font> is not affected when the <font style="font-style: italic;">dcmR</font> and <font style="font-style: italic;">dcmA</font> transcriptional units are placed on separate replicons thereby suggesting that their topology is independent of the regulatory network. It is therefore suggested that DcmR binds the DNA in the intergenic region with the simplest model of its mode of action being as a trans-acting DNA-binding repressor; however this remains to be fully validated [1].<br><br> |
| - | + | We have therefore proceeded on the assumption that DcmR is directly influenced by the presence or absence of DCM and furthermore that we can use <font style="font-style: italic;">dcmR</font>, the intergenic region and <font style="font-style: italic;">dcmA</font> alone to characterise the regulatory network. <br><br> | |
[1] La Roche, S. D., and T. Leisinger. "Identification of <font style="font-style: italic;">dcmR</font>, the regulatory gene governing expression of dichloromethane dehalogenase in Methylobacterium sp. strain DM4." Journal of bacteriology 173.21 (1991): 6714-6721. <br> | [1] La Roche, S. D., and T. Leisinger. "Identification of <font style="font-style: italic;">dcmR</font>, the regulatory gene governing expression of dichloromethane dehalogenase in Methylobacterium sp. strain DM4." Journal of bacteriology 173.21 (1991): 6714-6721. <br> | ||
| Line 101: | Line 104: | ||
<br><br> | <br><br> | ||
<h1>Characterising the DcmR - DCM - P_dcmA interaction</h1> | <h1>Characterising the DcmR - DCM - P_dcmA interaction</h1> | ||
| - | To find out whether the | + | To find out whether the DcmR acts as a repressor or an activator on the promoter of the <font style="font-style: italic;">dcmA</font> gene, we attempted to build the genetic circuit shown above on the right. Having <font style="font-style: italic;">dcmR</font> under inducible TetR expression should allow us to have very good control of the amount of DcmR present. Additionally a translational fusion with DcmR and a mCherry fluorescence tag will act as another confirmation to the amount of DcmR present. |
<br><br> | <br><br> | ||
We then extensively modelled the circuit to discover how the response of the system would differ if it was either of the two circuit systems. Click the modelling bubbles (pink) to find out exactly how we achieved this. | We then extensively modelled the circuit to discover how the response of the system would differ if it was either of the two circuit systems. Click the modelling bubbles (pink) to find out exactly how we achieved this. | ||
| Line 136: | Line 139: | ||
<h1>Predicting the mCherry fluorescence</h1> | <h1>Predicting the mCherry fluorescence</h1> | ||
| - | We simplified the first double repression by modelling it as an activation of <font style="font-style: italic;">dcmR</font> by ATC, albeit parameterised by different constants. This assumption is justified by the fact that we are able to precisely control the addition of ATC and | + | We simplified the first double repression by modelling it as an activation of <font style="font-style: italic;">dcmR</font> by anhydrous-tetracycline (ATC), albeit parameterised by different constants. This assumption is justified by the fact that we are able to precisely control the addition of ATC and measure the fluorescence of the mCherry. |
<br> | <br> | ||
<br> | <br> | ||
| Line 165: | Line 168: | ||
<br> | <br> | ||
| - | The differential equation that | + | The differential equation that describes this first step of the system is: |
| Line 179: | Line 182: | ||
<br><br><br><br><br><br> | <br><br><br><br><br><br> | ||
| - | Solving this ODE in Matlab (with zero basal transcription rate) predicts the following the response of the system: | + | Solving this ODE in Matlab (with a zero basal transcription rate) predicts the following the response of the system: |
<br><br> | <br><br> | ||
This model works assuming that sufficient TetR is always present. | This model works assuming that sufficient TetR is always present. | ||
| Line 195: | Line 198: | ||
As you can clearly see from the graph, the model predicts a large fluorescence increase as the input is added. This is the what we expect from the actual system and is the best approximation that is obtainable before we get experimental data. | As you can clearly see from the graph, the model predicts a large fluorescence increase as the input is added. This is the what we expect from the actual system and is the best approximation that is obtainable before we get experimental data. | ||
<br><br> | <br><br> | ||
| - | In the graph above, the model is set to have a basal transcription rate of zero. This is why there is a zero fluorescence response before the input has been added - this corresponds to the | + | In the graph above, the model is set to have a basal transcription rate of zero. This is why there is a zero fluorescence response before the input has been added - this corresponds to the Ptet promoter not being leaky. This basal rate will be calibrated alongside all of the other parameters in the model. |
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 277: | Line 256: | ||
<h1>Predicting the sfGFP fluorescence</h1> | <h1>Predicting the sfGFP fluorescence</h1> | ||
<h1>Introduction</h1> | <h1>Introduction</h1> | ||
| - | To allow us to characterize the second half of the genetic circuit, we needed to be able to predict the difference in response. To do this, we constructed models by cascading the differential equations according to the respective circuit structures thereby producing two different potential system responses. | + | To allow us to characterize the second half of the genetic circuit (DcmR regulating sfGFP), we needed to be able to predict the difference in response. To do this, we constructed models by cascading the differential equations according to the respective circuit structures thereby producing two different potential system responses. |
<br><br> | <br><br> | ||
We then set up the differential equations necessary to solve this problem in Matlab. The method and results are as detailed below: | We then set up the differential equations necessary to solve this problem in Matlab. The method and results are as detailed below: | ||
| Line 313: | Line 292: | ||
<div class="white_news_block2"> | <div class="white_news_block2"> | ||
| - | <a href="https://static.igem.org/mediawiki/2014/b/be/Oxford_Equations_explained.png"><img src="https://static.igem.org/mediawiki/2014/4/41/Oxford_equations.png" style="float:left;position:relative; width:50%;margin-right:25%;margin-left:25%;" /></a> | + | <a href="https://static.igem.org/mediawiki/2014/b/be/Oxford_Equations_explained.png" target="_blank"><img src="https://static.igem.org/mediawiki/2014/4/41/Oxford_equations.png" style="float:left;position:relative; width:50%;margin-right:25%;margin-left:25%;" /></a> |
<br><br> | <br><br> | ||
Oxford iGEM 2014 | Oxford iGEM 2014 | ||
| Line 338: | Line 317: | ||
Calculating the many parameters for this system will be undoubtedly challenging. | Calculating the many parameters for this system will be undoubtedly challenging. | ||
<br><br> | <br><br> | ||
| - | <a href="https://2014.igem.org/Team:Oxford/calculating_parameters">How are we calculating the parameters?</a> | + | <a href="https://2014.igem.org/Team:Oxford/calculating_parameters" target="_blank">How are we calculating the parameters?</a> |
<br><br> | <br><br> | ||
<a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show7">Go to the data section where we calculated parameters for this part of the circuit. | <a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show7">Go to the data section where we calculated parameters for this part of the circuit. | ||
| Line 353: | Line 332: | ||
| + | |||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <a href="#show10" class="show modelling-row" id="show10"><div class="modelling"> | ||
| + | <h1white>Stochastic modelling</h1white> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/4d/Oxford_plus-sign-clip-art.png" style="float:right;position:relative; width:2%;" /> | ||
| + | </div></a> | ||
| + | |||
| + | <a href="#hide10" class="hide" id="hide10"><div class="modelling"> | ||
| + | <h1white>Stochastic modelling</h1white></div></a> | ||
| + | <div class="list"> | ||
| + | <div class="white_news_block2"> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="white_news_block2"> | ||
| + | <h1>Stochastic Modelling</h1> | ||
| + | |||
| + | |||
| + | Stochastic modelling uses probability theory to predict the behaviour of a system. For our project, we used it to model the expression of the mCherry protein from bacteria. | ||
| + | |||
| + | <br><br> | ||
| + | We started with the Gillespie Algorithm, which considers the expression of mCherry to be binary; a molecule of mCherry is either expressed or degraded. Before we determined which event happened, we had to work out when the event happened. Using the random number r1 (taken from a uniform distribution between 0 and 1), we produced another random number τ, which determined the time until the next reaction. | ||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/8/89/Oxford_Matt_equations_1.jpg" style="float:left;position:relative; height:8%; width:20%;" /> | ||
| + | |||
| + | <br><br><br><br><br><br> | ||
| + | Where α0 represents the probability that any reaction will happen, given by the following equation: | ||
| + | <br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/5/5c/MattBoothEquation2.png" style="float:left;position:relative; height:4%; width:47%;" /> | ||
| + | <br><br><br><br><br><br> | ||
| + | |||
| + | We modelled the probability of a molecule of mCherry being created using the Michaelis-Menten model (α1), incorporating a basal transcription rate (b1). For the degradation, we assumed a simple proportional relationship: the more mCherry you have, the more likely it is that a molecule degrades (δ1). The constant of proportionality will be a function of the intrinsic life time of the protein in the cell. We considered there to be no ATC originally, then a large step in ATC at time=50. | ||
| + | <br><br> | ||
| + | To decide if mCherry was expressed, we looked at the percentage of events which were expressions. Then we compared this to a second random number r2 (again taken from a uniform distribution from 0 to 1). If the random number was lower, then mCherry was expressed. If it was higher, then a mCherry was degraded. In this way we make a weighted random choice about whether mCherry was expressed or degraded. We only stored the time and amount of mCherry when there was a event, to save on computation time. | ||
| + | |||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/8/86/MattBoothEquation3.png" style="float:left;position:relative; height:8%; width:30%;" /> | ||
| + | |||
| + | <br><br><br><br><br><br><br><br> | ||
| + | Stochastic modelling is useful because it can show us the stochastic effects which are often observed in individual bacteria. By calculating the variation of the mean of multiple mCherry producing bacteria, we can also work out the standard deviation. Then if we assume that the system varies with respect to the normal distribution, we can produce error bounds for the production of mCherry, such that we can say that 90% of the time we can expect the production of mCherry from a single bacterium to be within these two curves as seen in Figure 2. This could be useful for seeing if results are unexpected, or, if there are multiple outliers, that our model is incorrect. If we average an increasing number of bacteria, then the mean stochastic curve tends towards the deterministic response as seen in Figure 1. This is to be expected, as we are now looking at the system as a whole and fluctuations in the production from individual bacteria are averaged out. In terms of their use, when looking at small amounts of bacterium the stochastic model would be better, because real random fluctuations can be seen. For larger bacterial populations, the deterministic response models the growth very well. The stochastic model can also model large groups but requires large number of realisations which causes simulations to take a lot longer to run. | ||
| + | <br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/4/44/MattBoothStochasticNormal2.png" style="float:left;position:relative; height:18%; width:67.5%;" /> | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/9/9c/MattBoothIncreasing2.png" style="float:left;position:relative; height:18%; width:67.5%;" /> | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | When we originally ran the models, we picked arbitrary constants to view the general response. Later we created a code which was able to produce a line of best fit for fluorescence data from the lab; from this line the constants (a1,b1,d1,k1) could be determined. The exact calculations can be found on the "Calculating Parameters subsection". | ||
| + | <br><br> | ||
| + | |||
| + | However, because the of the size of expression term, the time between events are very small resulting in an almost deterministic response even with only 1 realisation as can be seen on Figure 3. | ||
| + | <br><br> | ||
| + | α1 = expression rate constant of dcmR = 16.5min-1 | ||
| + | <br> | ||
| + | k1 = Michaelis - Menten constant of dcmR = 0.015 ml/ng | ||
| + | <br> | ||
| + | d1 = degradation constant of dcmR = d1=0.00385min-1 | ||
| + | <br> | ||
| + | β1 = Basal transcription rate of dcmR = b1=1.81min-1 | ||
| + | <br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/b/b9/MattBoothStochasticData2.png" style="float:left;position:relative; height:18%; width:67.5%;" /> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| Line 388: | Line 454: | ||
<h1>Wetlab data showing response in level of mCherry expressed with different concs of ATC</h1> | <h1>Wetlab data showing response in level of mCherry expressed with different concs of ATC</h1> | ||
| - | By making a translational fusion of mCherry at the | + | By making a translational fusion of mCherry at the C terminus of the dcmR gene under the tet promoter and tet operator system (see our <a href="https://2014.igem.org/Team:Oxford/biosensor_construction">Construction page</a> for details) we could measure mCherry fluorescence to gain information about <font style="font-style: italic;">dcmR</font> induction by ATC. Expression was induced with various amounts of ATC and the following fluorescence data acquired. Exposure time was 0.2 seconds. As no calibration data was obtained using purified mCherry, the results have been left in fluorescence arbitrary units. Images were analysed using imageJ software.<br> |
mCherry fluorescence increases with amount of ATC used confirming that the dcmR gene was expressed under the control of the tet promoter and operator system.<br><br> | mCherry fluorescence increases with amount of ATC used confirming that the dcmR gene was expressed under the control of the tet promoter and operator system.<br><br> | ||
<img src="https://static.igem.org/mediawiki/parts/5/51/Oxford_DcmR-mCherry_expression_induced_by_0ng_ATC.png" | <img src="https://static.igem.org/mediawiki/parts/5/51/Oxford_DcmR-mCherry_expression_induced_by_0ng_ATC.png" | ||
style="float:left;position:relative; width:70%;" /> | style="float:left;position:relative; width:70%;" /> | ||
| - | <img src="https://static.igem.org/mediawiki/parts/1/19/Oxford_DcmR-mCherry_expression_induced_by_10ng_ATC.png" style="float: | + | <img src="https://static.igem.org/mediawiki/parts/1/19/Oxford_DcmR-mCherry_expression_induced_by_10ng_ATC.png" style="float:left;position:relative; width:70%;" /> |
<img src="https://static.igem.org/mediawiki/parts/6/63/Oxford_DcmR-mCherry_expression_induced_by_50ng_ATC.png" style="float:left;position:relative; width:70%;" /> | <img src="https://static.igem.org/mediawiki/parts/6/63/Oxford_DcmR-mCherry_expression_induced_by_50ng_ATC.png" style="float:left;position:relative; width:70%;" /> | ||
| - | <img src="https://static.igem.org/mediawiki/parts/c/cb/Oxford_DcmR-mCherry_expression_induced_by_100ng_ATC.png" style="float: | + | <img src="https://static.igem.org/mediawiki/parts/c/cb/Oxford_DcmR-mCherry_expression_induced_by_100ng_ATC.png" style="float:left;position:relative; width:70%;" /> |
<img src="https://static.igem.org/mediawiki/parts/3/37/Oxford_DcmR-mCherry_expression_induced_by_200ng_ATC.png" style="float:centre;position:relative; width:70%;" /> | <img src="https://static.igem.org/mediawiki/parts/3/37/Oxford_DcmR-mCherry_expression_induced_by_200ng_ATC.png" style="float:centre;position:relative; width:70%;" /> | ||
| - | <img src="https://static.igem.org/mediawiki/parts/e/e6/Oxford_Comparison_of_mean_flouresence_intensity_from_DcmR-mCherry_expression_induced_at_varying_amounts_of_ATC.png" style="float: | + | <img src="https://static.igem.org/mediawiki/parts/e/e6/Oxford_Comparison_of_mean_flouresence_intensity_from_DcmR-mCherry_expression_induced_at_varying_amounts_of_ATC.png" style="float:center;position:relative; width:70%;" /><br><br><br><br> |
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/e/e9/Microscopy_of_dcmR-mCherry_induction.png" style="float:center;position:relative; width:70%;margin-left:10%;" /> | ||
| + | <br><br>Figure 1. Epi-fluorescence microscopy images (green channel) of cells expressing dcmR-mCherry fusion induced by 0ng ATC, 50ng ATC,and 200ng (left to right) | ||
| Line 437: | Line 506: | ||
<h1>Introduction</h1> | <h1>Introduction</h1> | ||
| - | + | Our team were able to obtain good data for both the mCherry response of the system and the overall sfGFP response. This bubble shows how we adapted our model to make the most of the mCherry fluorescence data. | |
<br><br> | <br><br> | ||
The original data is shown on the right with error bars showing the standard error of the measurements. | The original data is shown on the right with error bars showing the standard error of the measurements. | ||
| Line 448: | Line 517: | ||
<a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show2">first modelling bubble</a> | <a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show2">first modelling bubble</a> | ||
on this page, shown here with a small basal rate (see | on this page, shown here with a small basal rate (see | ||
| - | <a href="https://static.igem.org/mediawiki/2014/b/be/Oxford_Equations_explained.png">where did these equations come from?</a> | + | <a href="https://static.igem.org/mediawiki/2014/b/be/Oxford_Equations_explained.png" target="_blank">where did these equations come from?</a> |
). This graph shows how the predicted fluorescence of the cells changes with time in response to an addition of ATC halfway along the time scale. At this stage, all input values, model parameters and therefore results are arbitrary. | ). This graph shows how the predicted fluorescence of the cells changes with time in response to an addition of ATC halfway along the time scale. At this stage, all input values, model parameters and therefore results are arbitrary. | ||
<br><br> | <br><br> | ||
<img src="https://static.igem.org/mediawiki/2014/4/46/Oxford_data2.png" style="float:right;position:relative; width:80%;margin-left:10%;margin-right:10%;" /> | <img src="https://static.igem.org/mediawiki/2014/4/46/Oxford_data2.png" style="float:right;position:relative; width:80%;margin-left:10%;margin-right:10%;" /> | ||
<br><br> | <br><br> | ||
| - | We then ran the model for the correct amount of time (2 hours 20mins incubation with ATC) and ran it for lots of different concentrations of ATC over the range that the wet-lab team | + | We then ran the model for the correct amount of time (2 hours 20mins incubation with ATC) and ran it for lots of different concentrations of ATC over the range that the wet-lab team tested. The parameters are still arbitrary at this point (the same as above) and the results of the graphs are therefore arbitrary are as well, but the input values are now correct. The graphic below shows how we used the existing model to obtain the same graphs as the wet-lab team had obtained. |
<br><br> | <br><br> | ||
The numerical inputs that were used to model this data set were therefore: | The numerical inputs that were used to model this data set were therefore: | ||
| Line 535: | Line 604: | ||
To find the parameters that make the model’s output match the data values, we turned to the code that we had developed for parameter calibration. | To find the parameters that make the model’s output match the data values, we turned to the code that we had developed for parameter calibration. | ||
<br><br> | <br><br> | ||
| - | <a href="https://2014.igem.org/Team:Oxford/calculating_parameters">How are we calculating the parameters?</a> | + | <a href="https://2014.igem.org/Team:Oxford/calculating_parameters" target="_blank">How are we calculating the parameters?</a> |
<br><br> | <br><br> | ||
This code gave the parameters as: | This code gave the parameters as: | ||
| Line 632: | Line 701: | ||
<div class="row"> | <div class="row"> | ||
<a href="#show6" class="show wetlab-row" id="show6"><div class="wetlab"> | <a href="#show6" class="show wetlab-row" id="show6"><div class="wetlab"> | ||
| - | <h1white> | + | <h1white>Is DcmR a repressor or an activator of the PdcmA/PdcmR promoters?</h1white> |
<img src="https://static.igem.org/mediawiki/2014/4/4d/Oxford_plus-sign-clip-art.png" style="float:right;position:relative; width:2%;" /> | <img src="https://static.igem.org/mediawiki/2014/4/4d/Oxford_plus-sign-clip-art.png" style="float:right;position:relative; width:2%;" /> | ||
</div></a> | </div></a> | ||
<a href="#hide6" class="hide" id="hide6"><div class="wetlab"> | <a href="#hide6" class="hide" id="hide6"><div class="wetlab"> | ||
| - | <h1white> | + | <h1white>Is DcmR a repressor or an activator of the PdcmA/PdcmR promoters?</h1white></div></a> |
<div class="list"> | <div class="list"> | ||
<div class="white_news_block2"> | <div class="white_news_block2"> | ||
| + | In order to evaluate the action of DcmR we measured sfGFP expression driven by the PdcmA promoter both in the presence of DcmR and not (whether the cell was transformed with our POXON-2 plasmid (BBa_K1446003) or not). The cells were grown in EZ rich media for 4hrs followed by fluorometry readings. The fluorescence results were then normalised to the OD readings of each sample to account for differences in cell density. | ||
| + | <br><br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/b/b7/Oxford_CharacGlen1.png" style="float:left;position:relative; width:80%;margin-bottom:2%;margin-left:10%;margin-right:10%;" /> | ||
| - | |||
| + | The results clearly show a decrease in sfGFP fluorescence upon addition and expression of dcmR. Therefore we can confidently conclude that DcmR acts as a repressor in both directions for the PdcmA and PdcmR promoter. | ||
<br><br> | <br><br> | ||
| + | <h1>Conclusion: DcmR is a repressor of both the PdcmA and PdcmR promoter.</h1> | ||
| - | |||
| - | |||
| - | The | + | |
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <a href="#show11" class="show wetlab-row" id="show11"><div class="wetlab"> | ||
| + | <h1white>DcmR at the bidirectional promoter? - A new tool for molecular biologists</h1white> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/4d/Oxford_plus-sign-clip-art.png" style="float:right;position:relative; width:2%;" /> | ||
| + | </div></a> | ||
| + | |||
| + | <a href="#hide11" class="hide" id="hide11"><div class="wetlab"> | ||
| + | <h1white>DcmR at the bidirectional promoter? - A new tool for molecular biologists</h1white></div></a> | ||
| + | <div class="list"> | ||
| + | <div class="white_news_block2"> | ||
| + | |||
| + | The expression profile of PdcmA and PdcmR was analysed. This was achieved using sfGFP in place of dcmA (for PdcmA studies) or dcmR (for PdcmR studies). To investigate the relative stoichiometries of expression between the two sides of this bidirectional promoter we ran a fluorescence time course experiment for 16 hours. The OD plot is seen below. We determined the ratio of GFP fluorescence for expression in the PdcmA direction and the PdcmR direction. The results are shown below: | ||
<br><br> | <br><br> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2014/7/7f/Oxford_charac3.png" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> | |
| + | <br> | ||
| + | <strong>Figure 2</strong>shows the the gene region under investigation and manipulation in the design of our biosensor. PdcmA and PdcmR describe the two directions of a bidirectional promoter that is bound and modulated by DcmR | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | We can see from these results that at both growth stages the relative expression is weighted towards PdcmR. An interesting result is that in the presence of DcmR this ratio is reduced such that expression through PdcmR is relatively increased while expression through PdcmA is relatively decreased. This result is observed as a decrease in the PdcmA/PdcmR expression ratio. From our results this appears more statistically relevant in stationary phase. The cut off for exponential/stationary phase was set at 600 minutes as seen in the above graph. | ||
<br><br> | <br><br> | ||
| - | < | + | <img src="https://static.igem.org/mediawiki/2014/1/18/Oxford_CharacGlen2.png" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> |
| - | < | + | <h1>Our new bidirectional promoter offers a new tool to molecular biologists. Our new intergenic region is able to modulate different stoichiometries between genes inserted either side of it in response to the presence or absence of DcmR (available in our BioBrick BBa_K1446003). The relative stoichiometry (PdcmA/PdcmR activation) of ~0.9 being reduced to ~0.3 upon DcmR addition. This remarkable property will be crucial to any molecular biologists or future iGEM teams wishing to investigate the effect of different stoichiometries on the dynamics of various systems. </h1> |
| + | </div> | ||
| - | + | </div> | |
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <a href="#show12" class="show wetlab-row" id="show12"><div class="wetlab"> | ||
| + | <h1white>What influence does DCM have on the action of DcmR?</h1white> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/4d/Oxford_plus-sign-clip-art.png" style="float:right;position:relative; width:2%;" /> | ||
| + | </div></a> | ||
| + | |||
| + | <a href="#hide12" class="hide" id="hide12"><div class="wetlab"> | ||
| + | <h1white>What influence does DCM have on the action of DcmR?</h1white></div></a> | ||
| + | <div class="list"> | ||
| + | <div class="white_news_block2"> | ||
| + | In order to determine the influence of DCM on our DcmR repression system we grew MG1655 cells transformed with either PdcmA or PdcmR with DcmR and either with or without DCM. The cells were grown for 4 hours with no DCM added to grow to mid-log phase. After this time half the cells were treated with 30mM DCM and grown for an additional 2 hours before fluorescence was measured. The results can be seen below with fluorescence normalised for OD(600): | ||
<br><br> | <br><br> | ||
| - | + | For the PdcmA promoter both with and without DcmR: | |
<br><br> | <br><br> | ||
| - | + | <img src="https://static.igem.org/mediawiki/2014/e/ec/Oxford_CharacGlen3.png" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> | |
| + | |||
| + | For the PdcmR promoter both with and without DcmR: | ||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/2/2c/Oxford_CharacGlen4.png" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> | ||
| + | |||
| + | From these results we can see that in the direction of PdcmA DCM has a modulating effect of relieving DcmR repression thus DCM acts through depression. Interestingly, DCM appears to decrease sfGFP fluorescence in the absence of DcmR. | ||
| + | In the direction of PdcmR DCM appears to have no significant effect on the expression of sfGFP. Therefore in the native system we suggest that DCM has a modulating effect through depression of dcmA expression. Additionally DCM shows no effect on the auto regulation of dcmR expression. | ||
| + | <br><br> | ||
| + | In the context of our biosensor this means that DCM is capable of modulating expression of sfGFP through interaction through the PdcmA repressor DcmR. | ||
| + | <br><br> | ||
| + | Ultimately we can conclude that DCM acts to encourage expression of sfGFP through derepression: consistent with our double repression model. | ||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/c/c0/Oxford_CharacGlen5.png" style="float:right;position:relative; width:60%; margin-right:20%;margin-bottom:2%;margin-left:20%;" /> | ||
| + | <br><br> | ||
| + | |||
| + | These images taken on an epi-fluorescence microscope (exposure time 0.4seconds) physically show the fluorescence from the same cells as used in the time course study as above. | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2014/1/19/Microscopy_comparison_-_A-E_%2B_and_-DCM.png" style="float:right;position:relative; width:60%; margin-right:20%;margin-bottom:2%;margin-left:20%;" /> | ||
| + | <br><br> | ||
| + | |||
| + | |||
| + | <h1>Discussion: | ||
| + | <br><br> | ||
| + | The results displayed here demonstrate the repression nature of DcmR on PdcmA and PdcmR bidirectional promoter. The modulation by DCM on the PdcmA promoter we have observed offers a constructive route towards a Dichloromethane biosensor. In this way we are able to use a reporter gene downstream of PdcmA to respond to the concentration of DCM. | ||
| + | <br><br> | ||
| + | Additionally the nature of the bidirectional promoter offers a uniquely useful tool to molecular biologists. The relative stoichiometry of two genes inserted either side of the bidirectional promoter can be can be modulated through the addition of DcmR (BBa_K1446003 - Oxford iGEM). This can be utilised easily for investigations into the effect of protein stoichiometries between two interacting proteins. </h1> | ||
| Line 679: | Line 862: | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <a href="#show9" class="show modelling-row" id="show9"><div class="modelling"> | ||
| + | <h1white>Engineering analysis of the sfGFP data</h1white> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/4d/Oxford_plus-sign-clip-art.png" style="float:right;position:relative; width:2%;" /> | ||
| + | </div></a> | ||
| + | |||
| + | <a href="#hide9" class="hide" id="hide9"><div class="modelling"> | ||
| + | <h1white>Engineering analysis of the sfGFP data</h1white></div></a> | ||
| + | <div class="list"> | ||
| + | <div class="white_news_block2"> | ||
| + | The data that the sfGFP fluorescence data that the wet-lab team obtained presented the engineering team with the exciting possibility of fully parameterising our model of the synthetic circuit and therefore having the ability to make very good approximations of the overall behaviour of our biosensor. | ||
| + | <br><br> | ||
| + | <h1>Calculating the parameters</h1> | ||
| + | To calculate the parameters for behaviour of the second half of the genetic circuit (DcmR regulating sfGFP), we used a similar approach to the method we used to find the parameters for the top half of the circuit. This involves taking key bits of the data and analysing the corresponding equation. | ||
| + | <br><br> | ||
| + | <h1>Degradation rate constant (δ2)</h1> | ||
| + | The degradation half-life of the GFP protein is well documented in the literature and has a value of approximately 24 hours (1440 mins) [1] . This is very high and is one of the reasons that we chose GFP to be our fluorescence indicator. From this value of the half life, it is possible to calculate the degradation rate constant using mathematics: | ||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/5/56/Oxford_CharacOV1.png" style="float:left;position:relative; width:15%; margin-right:85%;margin-bottom:2%;margin-left:0%;" /> | ||
| + | |||
| + | <h1>Basal transcription rate constant (β3)</h1> | ||
| + | To find the basal transcription rate constant of the second half of the system (DcmR regulating sfGFP), we analysed the data of the fluorescence of just the Pdcma and the sfGFP gene. | ||
| + | <br><br> | ||
| + | The data shown on the graphs below shows clearly that the fluorescence stops increasing when the bacteria stop growing in the log phase. This means that we can’t reliably use the data from the stationary phase to provide parameters. We have taken this point to be 500 minutes into the data measurement. | ||
| + | |||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/5/59/Oxford_CharacOV2.png" style="float:left;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> | ||
| + | |||
| + | In terms of the modelling equations, this system is equivalent to setting [DcmR] to zero and ensuring that for this scenario, we set β1 = 0 (basal transcription rate of DcmR) . The mathematics below shows how we used the normalised data to find the basal transcription rate of sfGFP. Note how we’re using equation with parameters α3, β3 and k3 because the wet-lab team were able to prove that DcmR acts as a repressor on PdcmA. | ||
| + | |||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/c/c0/Oxford_DCMR_Equations2.png" style="float:left;position:relative; width:35%; margin-right:65%;margin-bottom:2%;margin-left:0%;" /> | ||
| + | |||
| + | |||
| + | Using the analytical solution outlined above, we were then able to characterize the single unknown parameter in the system (β3 - the basal rate of transcription of sfGFP). By again utilizing our data fitting algorithm alongside our hypothetical value for the basal rate of transcription of sfGFP, β3 was calculated to be 4.6. | ||
| + | <br><br> | ||
| + | |||
| + | Now that we know two of the four parameters for the second half of our synthetic circuit (DcmR regulating sfGFP), we needed to calculate the final two parameters to complete our model. To do this, we analysed wet-lab data that showed the system in the presence of DcmR. This now means that the non linear term in our ODE is non zero and the analysis becomes | ||
| + | <br><br> | ||
| + | <h1>Expression rate constant (α3) and Michaelis - Menten constant (k3)</h1> | ||
| + | <br> | ||
| + | This data is shown here. Note the difference in the right hand plot reaffirming that DcmR acts as a repressor on PdcmA. | ||
| + | |||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/6/61/Oxford_3withDcmR.jpg" style="float:left;position:relative; width:100%; margin-right:0%;margin-bottom:2%;margin-left:0%;" /> | ||
| + | |||
| + | Similar parameter calculation from this data gives: | ||
| + | <br><br> | ||
| + | <h1>α3 = 0.95 min^-1</h1> | ||
| + | |||
| + | <h1>k3 = 0.065 ml/ng</h1> | ||
| + | |||
| + | <br><br><br><br> | ||
| + | |||
| + | <h1>Using the completed model</h1> | ||
| + | Entering these parameters into the model and comparing the response to data allowed us to analyse how effective our modelling had been. An example of how the model fits with some of the data that we used to derive the parameters is shown below: | ||
| + | <br><br> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/c/ca/Oxford_CharacOV4.png" style="float:left;position:relative; width:100%; margin-right:0%;margin-bottom:2%;margin-left:0%;" /> | ||
| + | |||
| + | |||
| + | More on the further use of this model is available in the <a href="https://2014.igem.org/Team:Oxford/biosensor_optimisation">optimisation</a> section. | ||
| + | |||
| + | <br><br> | ||
| + | <h1>Reference</h1> | ||
| + | [1] Andersen JB1, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria.(1998) Appl Environ Microbiol. 64(6):2240-6 | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
Latest revision as of 03:44, 18 October 2014



























 "
"








 Deterministic models are very powerful tools for synthetic biology. They describe the behaviour of the bacteria at the population level and use Ordinary Differential Equations (ODEs) to relate each activation and repression. By constructing a cascade of differential equations one can build a realistic model of the average behaviour of the system.
Deterministic models are very powerful tools for synthetic biology. They describe the behaviour of the bacteria at the population level and use Ordinary Differential Equations (ODEs) to relate each activation and repression. By constructing a cascade of differential equations one can build a realistic model of the average behaviour of the system.



 Oxford iGEM 2014
Oxford iGEM 2014


















 This simplifies the equation to:
This simplifies the equation to:
 As we want our model to accurately predict the fluorescence, we will substitute the fluorescence value in place of the [DcmR] and rearrange:
As we want our model to accurately predict the fluorescence, we will substitute the fluorescence value in place of the [DcmR] and rearrange:
 Substituting in the value for δ1 that we found above and the basal steady state fluorescence level from the data (471 to 3 s.f.) gives the basal transcription rate as:
Substituting in the value for δ1 that we found above and the basal steady state fluorescence level from the data (471 to 3 s.f.) gives the basal transcription rate as:

 alongside the correct inputs:
alongside the correct inputs:
 The graph below shows the model's predictions plotted in the same figure as the data points that the wet-lab team obtained for the system:
The graph below shows the model's predictions plotted in the same figure as the data points that the wet-lab team obtained for the system:
 Plotting the model's output as a by interpolating between the calculated values makes the graph clearer:
Plotting the model's output as a by interpolating between the calculated values makes the graph clearer:


 The results clearly show a decrease in sfGFP fluorescence upon addition and expression of dcmR. Therefore we can confidently conclude that DcmR acts as a repressor in both directions for the PdcmA and PdcmR promoter.
The results clearly show a decrease in sfGFP fluorescence upon addition and expression of dcmR. Therefore we can confidently conclude that DcmR acts as a repressor in both directions for the PdcmA and PdcmR promoter.

 For the PdcmR promoter both with and without DcmR:
For the PdcmR promoter both with and without DcmR:
 From these results we can see that in the direction of PdcmA DCM has a modulating effect of relieving DcmR repression thus DCM acts through depression. Interestingly, DCM appears to decrease sfGFP fluorescence in the absence of DcmR.
In the direction of PdcmR DCM appears to have no significant effect on the expression of sfGFP. Therefore in the native system we suggest that DCM has a modulating effect through depression of dcmA expression. Additionally DCM shows no effect on the auto regulation of dcmR expression.
From these results we can see that in the direction of PdcmA DCM has a modulating effect of relieving DcmR repression thus DCM acts through depression. Interestingly, DCM appears to decrease sfGFP fluorescence in the absence of DcmR.
In the direction of PdcmR DCM appears to have no significant effect on the expression of sfGFP. Therefore in the native system we suggest that DCM has a modulating effect through depression of dcmA expression. Additionally DCM shows no effect on the auto regulation of dcmR expression.



 In terms of the modelling equations, this system is equivalent to setting [DcmR] to zero and ensuring that for this scenario, we set β1 = 0 (basal transcription rate of DcmR) . The mathematics below shows how we used the normalised data to find the basal transcription rate of sfGFP. Note how we’re using equation with parameters α3, β3 and k3 because the wet-lab team were able to prove that DcmR acts as a repressor on PdcmA.
In terms of the modelling equations, this system is equivalent to setting [DcmR] to zero and ensuring that for this scenario, we set β1 = 0 (basal transcription rate of DcmR) . The mathematics below shows how we used the normalised data to find the basal transcription rate of sfGFP. Note how we’re using equation with parameters α3, β3 and k3 because the wet-lab team were able to prove that DcmR acts as a repressor on PdcmA.
 Using the analytical solution outlined above, we were then able to characterize the single unknown parameter in the system (β3 - the basal rate of transcription of sfGFP). By again utilizing our data fitting algorithm alongside our hypothetical value for the basal rate of transcription of sfGFP, β3 was calculated to be 4.6.
Using the analytical solution outlined above, we were then able to characterize the single unknown parameter in the system (β3 - the basal rate of transcription of sfGFP). By again utilizing our data fitting algorithm alongside our hypothetical value for the basal rate of transcription of sfGFP, β3 was calculated to be 4.6.
 Similar parameter calculation from this data gives:
Similar parameter calculation from this data gives:
 More on the further use of this model is available in the
More on the further use of this model is available in the 

































