Team:Lethbridge/results
From 2014.igem.org
| (22 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
</style> | </style> | ||
| - | + | <br> | |
| - | <center><a href="https://2014.igem.org/Team:Lethbridge/project"><image src="https://static.igem.org/mediawiki/2014/8/8f/Neuro_Results_Header.jpg" | + | <center><a href="https://2014.igem.org/Team:Lethbridge/project"><image src="https://static.igem.org/mediawiki/2014/8/8f/Neuro_Results_Header.jpg" width="600px" height="300px"/></a></center> |
| - | < | + | </html> |
| - | |||
<BLOCKQUOTE> | <BLOCKQUOTE> | ||
| Line 22: | Line 21: | ||
<h2>Team Parts Sandbox</h2> | <h2>Team Parts Sandbox</h2> | ||
<center> | <center> | ||
| - | <table border="0" style="background-color:#FFF" width=" | + | <table border="0" style="background-color:#FFF" width="100%" cellpadding="0" cellspacing="5"> |
<tr> | <tr> | ||
<td><strong>Name</strong></td> | <td><strong>Name</strong></td> | ||
| Line 41: | Line 40: | ||
<tr> | <tr> | ||
<td>[http://parts.igem.org/Part:BBa_K1419001 BBa_K1419001]</td> | <td>[http://parts.igem.org/Part:BBa_K1419001 BBa_K1419001]</td> | ||
| - | <td> | + | <td>Device</td> |
<td>Arabinose inducible lysis casette with RNA-IN</td> | <td>Arabinose inducible lysis casette with RNA-IN</td> | ||
<td>Zak Stinson</td> | <td>Zak Stinson</td> | ||
| - | <td> | + | <td>2982</td> |
<td> </td> | <td> </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>[http://parts.igem.org/Part:BBa_K1419002 BBa_K1419002]</td> | <td>[http://parts.igem.org/Part:BBa_K1419002 BBa_K1419002]</td> | ||
| - | <td> | + | <td>RNA Part</td> |
<td>RNA-OUT component for an Arabinose inducible lysis casette</td> | <td>RNA-OUT component for an Arabinose inducible lysis casette</td> | ||
<td>Harland Brandon</td> | <td>Harland Brandon</td> | ||
| - | <td>117 | + | <td>117</td> |
<td> </td> | <td> </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
<td>[http://parts.igem.org/Part:BBa_K1419003 BBa_K1419003]</td> | <td>[http://parts.igem.org/Part:BBa_K1419003 BBa_K1419003]</td> | ||
| - | <td> | + | <td>RNA Part</td> |
| - | <td>RNA- | + | <td>RNA-IN RBS Universal Adaptor for Translation Riboregulation</td> |
<td>Harland Brandon</td> | <td>Harland Brandon</td> | ||
| - | <td> | + | <td>39</td> |
<td> </td> | <td> </td> | ||
</tr> | </tr> | ||
| Line 68: | Line 67: | ||
<td>TEV Protease</td> | <td>TEV Protease</td> | ||
<td>Harland Brandon</td> | <td>Harland Brandon</td> | ||
| - | <td> | + | <td>740</td> |
<td> </td> | <td> </td> | ||
</tr> | </tr> | ||
| Line 76: | Line 75: | ||
<td>Lamp2B-Clover </td> | <td>Lamp2B-Clover </td> | ||
<td>Harland Brandon</td> | <td>Harland Brandon</td> | ||
| - | <td> | + | <td>2170</td> |
<td><center> Yes </center></td> | <td><center> Yes </center></td> | ||
</tr> | </tr> | ||
| Line 86: | Line 85: | ||
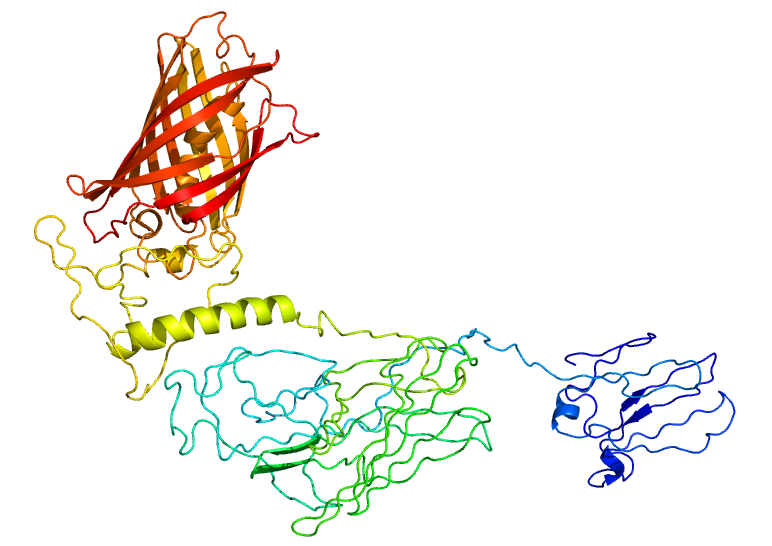
<p class="ex">To test if our RVG-Lamp2B construct was being appropriately expressed and folded, we fused Clover (a green fluorescent protein) to the C-terminus of Lamp2B. Using the Phyre2 server, we were able to generate a model of the predicted structure of our RVG-Lamp2B-Clover fusion protein based on homology and multi-template <i>ab initio</i> modeling.</p> | <p class="ex">To test if our RVG-Lamp2B construct was being appropriately expressed and folded, we fused Clover (a green fluorescent protein) to the C-terminus of Lamp2B. Using the Phyre2 server, we were able to generate a model of the predicted structure of our RVG-Lamp2B-Clover fusion protein based on homology and multi-template <i>ab initio</i> modeling.</p> | ||
| - | |||
| - | |||
<center>[[Image:Lamp2B_Clover_Structure.jpg|500px]]</center> | <center>[[Image:Lamp2B_Clover_Structure.jpg|500px]]</center> | ||
| Line 95: | Line 92: | ||
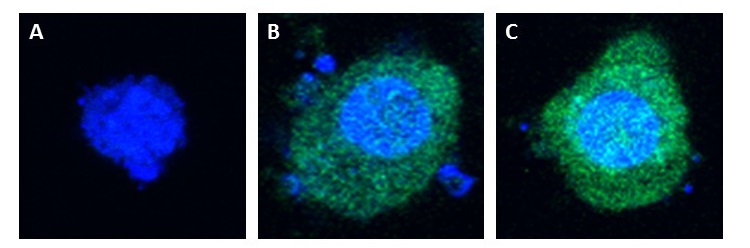
<center>[[Image:Lamp2B_Clover_exosomes.JPG|700px]]</center> | <center>[[Image:Lamp2B_Clover_exosomes.JPG|700px]]</center> | ||
| - | <p class="ex"><b>Figure 2. Confocal (60x) images of non-lipofected control HEK-293 cells versus pcDNA3.0-RVG-Lamp2B-Clover and pcDNA3.0-Clover lipofected HEK-293 cells.</b> Nuclei were counterstained with DAPI. A | + | <p class="ex"><b>Figure 2. Confocal (60x) images of non-lipofected control HEK-293 cells versus pcDNA3.0-RVG-Lamp2B-Clover and pcDNA3.0-Clover lipofected HEK-293 cells.</b> Nuclei were counterstained with DAPI (blue). A) Non-lipofected control HEK-293 cells. B) HEK-293 cells expressing Clover (green). C) Hek-293 cells expressing RVG-Lamp2B-Clover (green).</p><br> |
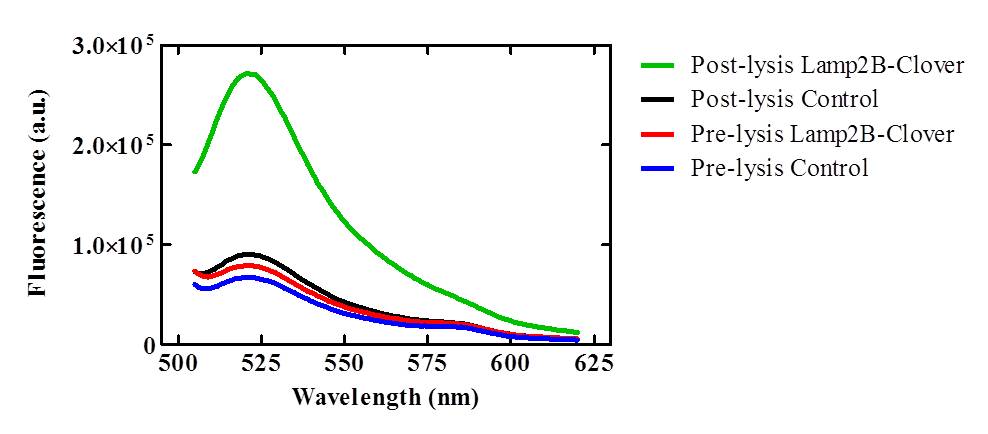
| - | <p class="ex">To confirm that the RVG-Lamp2B-Clover protein construct was being targeted to exosomal membranes (and were not simply cytosolic), culture media was collected from the lipofected and control HEK-293 cell cultures and exosomes were purified via ultracentrifugation. After lysis, the exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected cultures demonstrated an elevated level of fluorescence (505- | + | <p class="ex">To confirm that the RVG-Lamp2B-Clover protein construct was being targeted to exosomal membranes (and were not simply cytosolic), culture media was collected from the lipofected and control HEK-293 cell cultures and exosomes were purified via ultracentrifugation. After lysis, the exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected cultures demonstrated an elevated level of fluorescence (505-620 nm) that corresponds to the emission wavelength of Clover (515 nm) indicating appropriate targeting of the engineered protein to exosomal membranes.</p> |
<center>[[Image:Fluorimeter_data.jpg|700px]]</center> | <center>[[Image:Fluorimeter_data.jpg|700px]]</center> | ||
| - | <p class="ex"><b>Figure 3. Exosomes were isolated from HEK-293 cells expressing the RVG-Lamp2B-Clover construct and lysed with 0.5% (w/v) sodium deoxycholate | + | <p class="ex"><b>Figure 3. Lysed exosomes demonstrate elevated fluorescence in the 505-620 nm range relative to control and unlysed samples.</b> Clover has an emission wavelength of 515 nm. Exosomes were isolated from HEK-293 cells expressing the RVG-Lamp2B-Clover construct and lysed with 0.5% (w/v) sodium deoxycholate.</p><br> |
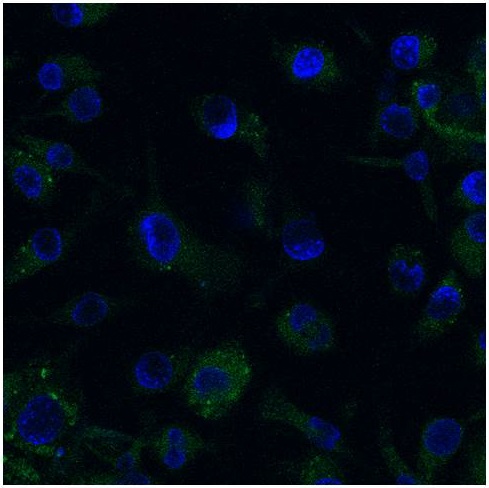
| - | <p class="ex"> | + | <p class="ex">Mouse microglia (EOC 13.31) cell cultures were subsequently transfected with the RVG-Lamp2B-Clover construct and demonstrated an elevation in Clover expression similar to that of lipofected HEK-293 cells.</p> |
| + | <center>[[Image:Microglia_Clover.JPG|400px]]</center> | ||
| + | <br> | ||
| + | <p class="ex"><b>Figure 4. Confocal (60x) image of pcDNA3.0-RVG-Lamp2B-Clover lipofected EOC 13.31 (mouse microglia) cells.</b> Nuclei were counterstained with DAPI (blue) and extranuclear Clover expression is evident in green.</p><br> | ||
| + | <html><p class="ex">We are now in the process of replacing Clover with a Gal4 DNA-binding protein (<a href="http://parts.igem.org/Part:BBa_K105007">K105007</a>) to the C-terminus of Lamp2B to anchor the therapeutic NeuroD1 plasmid within newly formed exosomes.</p></html> | ||
<h2>Astrocytic expression of <i>NeuroD1</i> and resulting phenotype</h2> | <h2>Astrocytic expression of <i>NeuroD1</i> and resulting phenotype</h2> | ||
| - | <p class="ex">To test if expression of NeuroD1 | + | <p class="ex">To test if expression of NeuroD1 had a phenotypic effect on astrocytes, pcDNA3.0-NeuroD1-Clover was transfected into C8-D30 cultures. After one week, both control and transfected astrocyte cells demonstrated extensive cytosolic Doublecortin (DCX) expression (an indicator of immature neurons) and very weak nuclear expression of NeuN (an indicator of mature neurons). Furthermore, the cell bodies and processes were still astrocytic in appearance and no Clover expression was evident in transfected cultures, indicating either a problem with the construction or expression of the plasmid and/or that the cell line is not representative of typical mature astrocytes. We are currently addressing both of these possibilities.</p> |
| + | |||
<center>[[Image:Astrocyte_Clover_NeuN.JPG|700px]]</center> | <center>[[Image:Astrocyte_Clover_NeuN.JPG|700px]]</center> | ||
| - | <p class="ex"><b>Figure | + | |
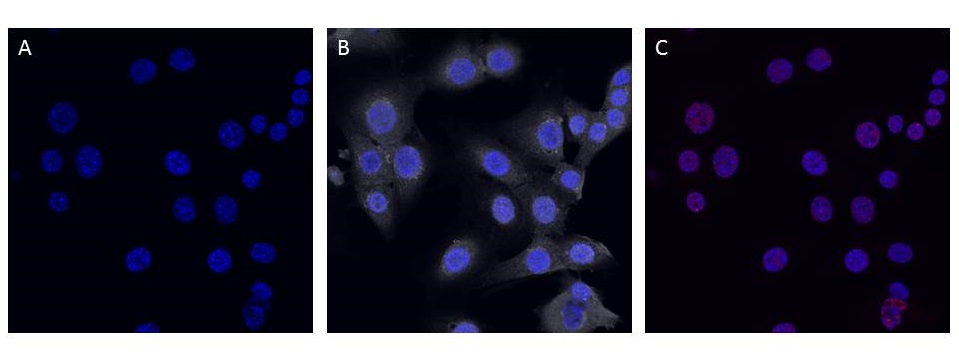
| + | <p class="ex"><b>Figure 5. Confocal (60x) image of C8-D30 (mouse astrocyte) cells transfected with pcDNA3.0-NeuroD1-Clover and non-transfected controls (not shown) both demonstrate elevated cytosolic Doublecortin labeling and weak nuclear NeuN expression.</b> Nuclei were counterstained with DAPI (blue). A) No Clover expression (green) was evident in transfected astrocytes. B) Extensive cytosolic Doublecortin (grey) staining was observed in both controls (not shown) and transfected cells. C) Weak nuclear NeuN (red) labeling was evident in both controls (not shown) and transfected cells. | ||
| + | </p><br> | ||
| + | |||
| + | <h2>Exosome Purification</h2> | ||
| + | |||
| + | <center>[[Image:UofLInset.PNG|700px]]</center> | ||
| + | |||
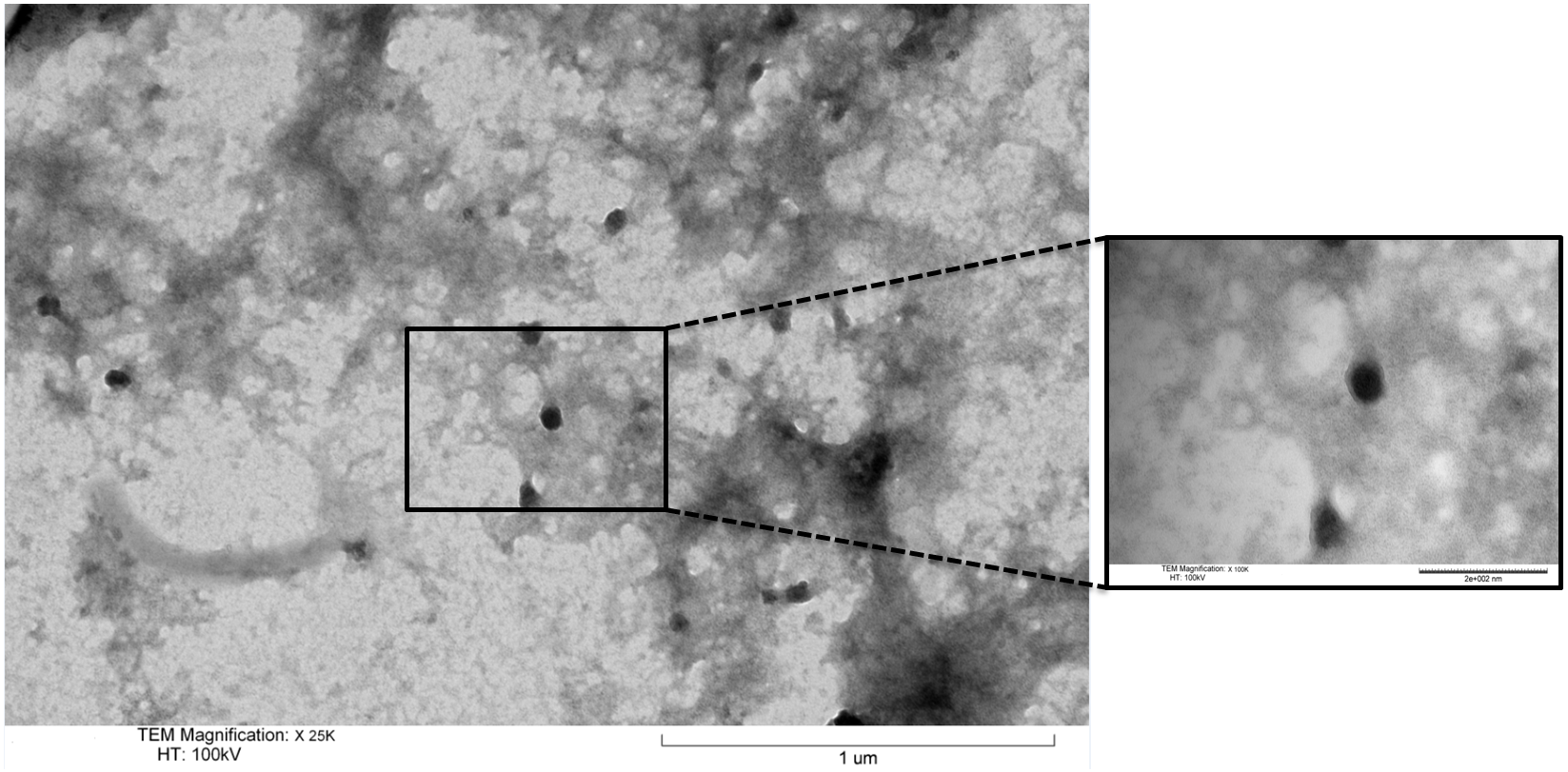
| + | <p class="ex"><b>Figure 6. Transmission Electron Microscopy micrograph of isolated exosomes.</b> These were fixed in 2% PFA and negatively stained with uranyl acetate. The size and distinctive morphology of the stained exosome isolates conforms with previous reports in the literature. </p><br> | ||
Latest revision as of 03:57, 18 October 2014

Contents
Results
Team Parts Sandbox
Name Type Description Designer Length Favorite Part [http://parts.igem.org/Part:BBa_K1419000 BBa_K1419000] Device Arabinose inducible lysis casette Zak Stinson 3002 [http://parts.igem.org/Part:BBa_K1419001 BBa_K1419001] Device Arabinose inducible lysis casette with RNA-IN Zak Stinson 2982 [http://parts.igem.org/Part:BBa_K1419002 BBa_K1419002] RNA Part RNA-OUT component for an Arabinose inducible lysis casette Harland Brandon 117 [http://parts.igem.org/Part:BBa_K1419003 BBa_K1419003] RNA Part RNA-IN RBS Universal Adaptor for Translation Riboregulation Harland Brandon 39 [http://parts.igem.org/Part:BBa_K1419004 BBa_K1419004] Device TEV Protease Harland Brandon 740 [http://parts.igem.org/Part:BBa_K1419005 BBa_K1419005] Device Lamp2B-Clover Harland Brandon 2170 Yes Expression and localization of RVG-Lamp2B protein construct
To test if our RVG-Lamp2B construct was being appropriately expressed and folded, we fused Clover (a green fluorescent protein) to the C-terminus of Lamp2B. Using the Phyre2 server, we were able to generate a model of the predicted structure of our RVG-Lamp2B-Clover fusion protein based on homology and multi-template ab initio modeling.
Figure 1. The predicted structure of the RVG-Lamp2B-Clover fusion protein. The Clover domain is shown in red with Lamp2B below in green and the RVG domain evident at the bottom in blue.
The pcDNA3.0-RVG-Lamp2B-Clover plasmid and a pcDNA3.0-Clover control were lipofected into separate Human Embryonic Kidney (HEK-293) cell cultures (HEK-293 cells are a resilient and rapidly dividing cell line also known to produce exosomes). In both lipofected cultures, there was elevated cytosolic green fluorescence relative to non-lipofected controls, which confirms expression and proper folding of the protein construct.
Figure 2. Confocal (60x) images of non-lipofected control HEK-293 cells versus pcDNA3.0-RVG-Lamp2B-Clover and pcDNA3.0-Clover lipofected HEK-293 cells. Nuclei were counterstained with DAPI (blue). A) Non-lipofected control HEK-293 cells. B) HEK-293 cells expressing Clover (green). C) Hek-293 cells expressing RVG-Lamp2B-Clover (green).
To confirm that the RVG-Lamp2B-Clover protein construct was being targeted to exosomal membranes (and were not simply cytosolic), culture media was collected from the lipofected and control HEK-293 cell cultures and exosomes were purified via ultracentrifugation. After lysis, the exosomes isolated from pcDNA3.0-RVG-Lamp2B-Clover lipofected cultures demonstrated an elevated level of fluorescence (505-620 nm) that corresponds to the emission wavelength of Clover (515 nm) indicating appropriate targeting of the engineered protein to exosomal membranes.
Figure 3. Lysed exosomes demonstrate elevated fluorescence in the 505-620 nm range relative to control and unlysed samples. Clover has an emission wavelength of 515 nm. Exosomes were isolated from HEK-293 cells expressing the RVG-Lamp2B-Clover construct and lysed with 0.5% (w/v) sodium deoxycholate.
Mouse microglia (EOC 13.31) cell cultures were subsequently transfected with the RVG-Lamp2B-Clover construct and demonstrated an elevation in Clover expression similar to that of lipofected HEK-293 cells.
Figure 4. Confocal (60x) image of pcDNA3.0-RVG-Lamp2B-Clover lipofected EOC 13.31 (mouse microglia) cells. Nuclei were counterstained with DAPI (blue) and extranuclear Clover expression is evident in green.
We are now in the process of replacing Clover with a Gal4 DNA-binding protein (K105007) to the C-terminus of Lamp2B to anchor the therapeutic NeuroD1 plasmid within newly formed exosomes.
Astrocytic expression of NeuroD1 and resulting phenotype
To test if expression of NeuroD1 had a phenotypic effect on astrocytes, pcDNA3.0-NeuroD1-Clover was transfected into C8-D30 cultures. After one week, both control and transfected astrocyte cells demonstrated extensive cytosolic Doublecortin (DCX) expression (an indicator of immature neurons) and very weak nuclear expression of NeuN (an indicator of mature neurons). Furthermore, the cell bodies and processes were still astrocytic in appearance and no Clover expression was evident in transfected cultures, indicating either a problem with the construction or expression of the plasmid and/or that the cell line is not representative of typical mature astrocytes. We are currently addressing both of these possibilities.
Figure 5. Confocal (60x) image of C8-D30 (mouse astrocyte) cells transfected with pcDNA3.0-NeuroD1-Clover and non-transfected controls (not shown) both demonstrate elevated cytosolic Doublecortin labeling and weak nuclear NeuN expression. Nuclei were counterstained with DAPI (blue). A) No Clover expression (green) was evident in transfected astrocytes. B) Extensive cytosolic Doublecortin (grey) staining was observed in both controls (not shown) and transfected cells. C) Weak nuclear NeuN (red) labeling was evident in both controls (not shown) and transfected cells.
Exosome Purification
Figure 6. Transmission Electron Microscopy micrograph of isolated exosomes. These were fixed in 2% PFA and negatively stained with uranyl acetate. The size and distinctive morphology of the stained exosome isolates conforms with previous reports in the literature.
 "
"