Team:Imperial/Gluconacetobacter
From 2014.igem.org
| (80 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
<li><a data-scroll href="#aim">Aims</a> | <li><a data-scroll href="#aim">Aims</a> | ||
</li> | </li> | ||
| - | + | <li><a data-scroll href="#genome_sequencing">Genome Sequencing</a> | |
</li> | </li> | ||
| - | + | <li><a data-scroll href="#toolbox"><em>G. xylinus</em> toolbox</a> | |
</li> | </li> | ||
| - | + | <li><a data-scroll href="#references">References</a> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</li> | </li> | ||
</ul> | </ul> | ||
| Line 32: | Line 28: | ||
<div class="pure-u-1-2"> | <div class="pure-u-1-2"> | ||
<h2>Overview</h2> | <h2>Overview</h2> | ||
| - | <p>Bacterial cellulose has great potential in many areas, including water purification, tissue scaffolds, wound dressings, etc., however, until now, all bacterial cellulose-based materials have been created using chemical or physical post-production processing, not genetic engineering. This is due to the lack of well-developed tools and methods for <em>Gluconacetobacter</em> genetic engineering, as well as the lack of genome sequence of the highest cellulose-producing strain ATCC 53582. We have overcome the numerous difficulties associated with <em>G.xylinus</em> genetic engineering, and turned <em>G.xylinus | + | <p>Bacterial cellulose has great potential in many areas, including water purification, tissue scaffolds, wound dressings, etc., however, until now, all bacterial cellulose-based materials have been created using chemical or physical post-production processing, not genetic engineering. This is due to the lack of well-developed tools and methods for <em>Gluconacetobacter</em> genetic engineering, as well as the lack of genome sequence of the highest cellulose-producing strain ATCC 53582. We have overcome the numerous difficulties associated with <em>G.xylinus</em> genetic engineering, and turned <em>G.xylinus igem</em> and ATCC 53852 strains into new platforms for the production of cellulose-based biomaterials by sequencing the genomes of ATCC 53582 and a new strain G. xylinus igem, creating a genetic toolbox of consisting of five new plasmid backbones and around 40 widely used genes, and developing a set of new and improved protocols for <em>G.xylinus</em> genetic engineering. </p> |
</div> | </div> | ||
<div class="pure-u-1-2"> | <div class="pure-u-1-2"> | ||
<h2>Key Achievements </h2> | <h2>Key Achievements </h2> | ||
<ul> | <ul> | ||
| - | <li>Isolated a new strain of <em>Gluconacetobacter</em> from Kombucha tea and characterized its properties fully.</li> | + | <li>Isolated a new strain of <em>Gluconacetobacter</em> (named <em>G. xylinus</em> igem) from Kombucha tea and characterized its properties fully.</li> |
| - | <li>Sequenced the previously unknown genomes of | + | <li>Sequenced the previously unknown genomes of <em>G. xylinus</em> ATCC 53582 and <em>G. xylinus</em> igem strains - the first genomes sequenced in the history of iGEM </li> |
| - | <li>Discovered four new plasmids capable of replication in <em>Gluconacetobacter</em> species - pSEVA321, pSEVA331, pSEVA351 and pBAV1K, which replicate both in <em>G. xylinus</em> ATCC 53582 and in | + | <li>Discovered four new plasmids capable of replication in <em>Gluconacetobacter</em> species - pSEVA321, pSEVA331, pSEVA351 and pBAV1K, which replicate both in <em>G. xylinus</em> ATCC 53582 and igem strains as well as in <em>E. coli</em>.</li> |
| - | <li> | + | <li>Were the first in science to create transgenic cells of <em>G.xylinus</em> igem strain. </li> |
| - | <li>Using our discovered plasmids, created a genetic toolbox consisting of 40 genes for <em>G. xylinus</em> engineering and expressed them in the ATCC 53582 and | + | <li>Using our discovered plasmids, created a genetic toolbox consisting of 40 genes for <em>G. xylinus</em> engineering and expressed them in the ATCC 53582 and igem.</li> |
| - | <li>Developed a set of new and improved protocols for efficient genetic engineering of <em>G. xylinus</em> | + | <li>Developed a set of new and improved protocols for efficient genetic engineering of <em>G. xylinus</em>.</li> |
| - | <li>In summary, | + | <li>In summary, turned <em>G. xylinus</em> ATCC 53582 and igem strains into new model organisms and developed the necessary tools to create a platform for genetic engineering of new cellulose-based biomaterials.</li> |
| Line 58: | Line 54: | ||
</section> | </section> | ||
<section id="introduction"> | <section id="introduction"> | ||
| + | <br/> | ||
| + | <figure class="content-image image-full"> | ||
| + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/f/f3/IC14_IMG_4132_RFP_glow.JPG"> | ||
| + | <figcaption> Genetically engineered <em>Gluconacetobacter xylinus</em> expressing RFP (top three) compared to wild-type (bottom). </figcaption> | ||
| + | </figure> | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
| + | |||
| + | |||
<p>Due to its high productivity, <em>G. xylinus</em> is the main species used for the production of bacterial cellulose based products. These range from superior wound dressings, artificial blood vessels, scaffolds for tissue engineering, high-quality speaker membranes, stronger paper, nata de coco and many others (Keshk 2014) .</p> | <p>Due to its high productivity, <em>G. xylinus</em> is the main species used for the production of bacterial cellulose based products. These range from superior wound dressings, artificial blood vessels, scaffolds for tissue engineering, high-quality speaker membranes, stronger paper, nata de coco and many others (Keshk 2014) .</p> | ||
<p><em>G. xylinus</em> has been the subject of the majority of studies into production of bacterial cellulose. However, research into <em>G. xylinus</em> has primarily focused on the effects of different culture conditions (such as composition of growth media, aeration and agitation) on cellulose productivity, productivity of different strains and different post-processing methods. Few attempts have been made to improve cellulose productivity or the physical properties of cellulose through genetic engineering (but see Chien 2006). Consequently most of the genetic engineering methods and tools available for model organisms, such as <em>E.coli</em>, have not been developed for <em>G. xylinus</em>. These tools must first be developed, in order to begin serious efforts to genetically engineer <em>G. xylinus</em> strains capable of producing novel biomaterials, more cellulose, and at a lower cost. | <p><em>G. xylinus</em> has been the subject of the majority of studies into production of bacterial cellulose. However, research into <em>G. xylinus</em> has primarily focused on the effects of different culture conditions (such as composition of growth media, aeration and agitation) on cellulose productivity, productivity of different strains and different post-processing methods. Few attempts have been made to improve cellulose productivity or the physical properties of cellulose through genetic engineering (but see Chien 2006). Consequently most of the genetic engineering methods and tools available for model organisms, such as <em>E.coli</em>, have not been developed for <em>G. xylinus</em>. These tools must first be developed, in order to begin serious efforts to genetically engineer <em>G. xylinus</em> strains capable of producing novel biomaterials, more cellulose, and at a lower cost. | ||
| + | In the middle of the summer, we isolated a new strain of Gluconacetobacter (named G. xylinus igem, which we previously called G. xylinus KI for Kombucha isolated ) from Kombucha tea and characterized its properties fully. We found it to be more amenable for genetic engineering due to higher transformation efficiencies than <em>G. xylinus</em> ATCC 53582 strain (see below), so we continued work on this strain, including sequencing its genome due to potential economic importance (it is a key component in the popular Kombucha tea). | ||
| + | |||
</p> | </p> | ||
| Line 69: | Line 74: | ||
<p>In addition to the lack of tools, the continuous cellulose production of <em>G. xylinus</em> introduces further problems for genetic engineering, as it results in a low growth rate (the division time of <em>G. xylinus</em> is 4 hours, which is 8 times slower than that of <em>E. coli</em>), formation of spontaneous cellulose non-producing mutants in agitated culture (detrimental for the engineering of high-producing strains) and difficulties in performing procedures such as transformation, plasmid DNA extraction, etc. due to the physically interfering cellulose pellicle.</p> | <p>In addition to the lack of tools, the continuous cellulose production of <em>G. xylinus</em> introduces further problems for genetic engineering, as it results in a low growth rate (the division time of <em>G. xylinus</em> is 4 hours, which is 8 times slower than that of <em>E. coli</em>), formation of spontaneous cellulose non-producing mutants in agitated culture (detrimental for the engineering of high-producing strains) and difficulties in performing procedures such as transformation, plasmid DNA extraction, etc. due to the physically interfering cellulose pellicle.</p> | ||
<p>Furthermore, although the highest cellulose-producing strain <em>G. xylinus</em> ATCC 53582 has been used in several studies, the genome sequence of this strain is still unknown, making it impossible to carry out targeted engineering of chromosomal genes, which is vital to achieve increased productivity.</p> | <p>Furthermore, although the highest cellulose-producing strain <em>G. xylinus</em> ATCC 53582 has been used in several studies, the genome sequence of this strain is still unknown, making it impossible to carry out targeted engineering of chromosomal genes, which is vital to achieve increased productivity.</p> | ||
| - | <p>We aim to solve all of these problems, by completing three major projects: sequencing the genomes of ATCC53582 and | + | <p>We aim to solve all of these problems, by completing three major projects: sequencing the genomes of ATCC53582 and igem strains and creating a large toolbox for <em>G. xylinus</em> genetic. We then aim to use these tools to increase and control cellulose productivity and create new biomaterials with wide-ranging properties, by incorporating proteins with different functions into the cellulose matrix (see <a href="https://2014.igem.org/Team:Imperial/Functionalisation">Functionalisation</a>).</p> |
</section> | </section> | ||
| - | <section id=" | + | <section id="genome_sequencing"> |
<h2>Genome sequencing</h2> | <h2>Genome sequencing</h2> | ||
| - | + | <figure class="content-image image-left"> | |
| - | + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/2/23/IC14_Alignment_example_output_file_2.jpg"> | |
| - | + | <figcaption> Figure1. Example of alignment of reads from genome sequencing against Komagataeibacter xylinusE25 genome. Alignment done using Tablet genome browser. </figcaption> | |
| - | + | </figure> | |
| - | + | <p>Although genome sequencing has become widely available due to the improvements in next-generation sequencing, even with smaller bacterial genomes (the genome of G. xylinus is approximately 3.5Mbp in size), genome sequencing is still unavailable for small to mid-sized research groups due to high costs. We performed a survey of prices offered by all commercially available service providers using the Illumina MiSeq next-generation sequencing platform (which is the most cost-effective method for bacterial genome sequencing) in Europe, and found that the average costs of fully sequencing a 3.5Mbp genome (which includes library preparation, sequencing, bioinformatics and gap filling) is approximately £3000 for "simple" to sequence genomes, but can far exceed that for repeat-rich, GC-rich or otherwise problematic genomes. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | In order to be able to sequence the genome with the limited budget available to our team, we did not use a commercial sequencing service, and decided to perform the full sequencing cycle ourselves, despite having no previous experience in genome sequencing, and were able to sequence two genomes for less than £1000. We created the gDNA library using Illumina Nextera kit, by modifying the original Illumina protocol to be amenable for small sample number (see gDNA library preparation protocol). During gDNA library preparation, we discovered that cellulose production of <em> G.xylinus </em> resulted in the presence of a persistent contaminant in DNA preparation, which persisted even after four consecutive DNA purification methods. This contamination interfered with effective library preparation, retarding the project several weeks, and ultimately prompted us to create a new and specific protocol for G.xylinus DNA extraction (see G.xylinus DNA extraction). We performed the sequencing run (multiplexed sequencing of ATCC 53582 and igem in a single run, paired-end 250bp reads using standard flow cell) on an Illumina MiSeq at Imperial College’s Genomics Laboratory. Despite problems with gDNA library preparation, the sequencing yielded good results (9,063,619 reads with 98.71% reads identified; see Figure 1 for an example output of read alignment against Komagateibacter xylinus E25 reference genome). We performed the quality control of the reads using FasQC and Trim Galore and completed the genome assembly with the assistance of Imperial Bioinformatics Support service using their in-house assembly pipeline BugBuilder. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p> | |
| - | + | As a result, we are proud to present here the first genomes sequenced in iGEM. The genome of <em>G. xylinus igem </em> strain is approximately 3.4Mbp in size, with a GC content of 63.52% (see Figure 2). Autoannotation using Prokka (Seemann 2014) identified in total 3052 putative coding sequences, 59 tRNA sequences and 24 rRNA and other RNA sequences. The genome <em> G. xylinus</em> ATCC53582 is approximately 2.8Mbp in size, with a GC content of 59.82% (see Figure 3). Autoannotation using Prokka (Seemann 2014) identified in total 2491 putative coding sequences, 46 tRNA sequences and 17 rRNA and other RNA sequences. We are currently performing on the comparitive genomics of these genomes and related genomes, and will properly publish our results in a peer-reviewed journal. The full genome sequences, as well as detailed information about <em>G. xylinus</em> igem strain and genome can be found on the Registry as part <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1321306">BBa_K1321306</a> | |
| + | and <em>G. xylinus</em> ATCC53582 as part <a href="http://parts.igem.org/wiki/index.php?title=Part:BBa_K1321305. "> BBa_K1321305 </a> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <figure class="content-image image-left"> | ||
| + | <img class="image-left" src="https://static.igem.org/mediawiki/2014/f/f5/IC14_G.xylinus_ATCC_53582_genome_overview.jpg"> | ||
| + | <figcaption> Figure 2. Overview of the genome of G.xylinus ATCC 53582. The genome is approximately 2.8Mbp in size, with a GC content of around 60% (internal circle) and approximately 2500 coding sequences (external circle; blue bands denote single CDSs). Note that this overview should be taken as a guide, as further analysis may reveal a more accurate organization of the contigs. </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | <figure class="content-image image-left"> | ||
| + | <img class="image-left" src="https://static.igem.org/mediawiki/2014/9/9b/IC14_G.xylinus_igem_genome_view.png"> | ||
| + | <figcaption> Figure 3. Overview of the genome of G.xylinus igem. The genome is approximately 3.4Mbp in size, with a GC content of around 63% (internal circle) and approximately 3000 coding sequences (external circle; blue bands denote single CDSs. Note that this overview should be taken as a guide, as further analysis may reveal a more accurate organization of the contigs. </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | </p> | ||
| + | <p> | ||
| + | </p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</section> | </section> | ||
| - | <section id=" | + | <section id="toolbox"> |
| - | <h2>Creating a genetic toolbox for G.xylinus</h2> | + | <h2>Creating a genetic toolbox for <em>G. xylinus</em></h2> |
| - | <p> | + | <p>Despite the great potential <em>G. xylinus</em> holds for new biomaterial production, very few tools for genetic engineering had been developed so far. Thus, in parallel to sequencing the genomes, we began building a new system in order to equip G.xylinus with all of the necessary tools and to turn the ATCC 53582 and igem strains into systems for genetically engineered biomaterials production. |
| + | We first characterized ATCC 53582 and igem strains, and created a set of new and improved protocols necessary for their genetic engineering (see <a href="https://2014.igem.org/Team:Imperial/Protocols">G.xylinus protocols</a>). As only a few vectors are available for G. xylinus genetic engineering (this is due to various reasons including patents, materials-transfer agreements, and retired or deceased researchers) we first created a set of new biobrick-compatible plasmid backbones capable of replication both in Gluconacetobacter and E.coli. In order to create the genetic toolkit, we decided to test out and modify already existing Biobricks in the parts registry (rather than creating a set of new parts) in order to retain the already existing familiarity genetic engineers have with these genes and make the toolkit more user-friendly. None of these parts had been used in Gluconacetobacter before however, so it was necessary to determine if they can be used in G.xylinus in the first place. Finally, we used this toolkit to clone Vitreoscilla hemoglobin gene Vhb (which is known to increase cellulose productivity and growth rate of G.xylinus (Chien et al. 2006)) behind a strong Anderson promoter, and express it in G.xylinus ATCC 53582 and G.xylinus igem strains to increase cellulose productivity and decrease its cost. In total, by cloning widely used parts into our <em>G. xylinus</em> shuttle vectors we created a toolkit consisting of: | ||
| + | <ul> | ||
| + | <li> 5 new shuttle vectors </li> | ||
| + | <li> 15 Anderson promoters </li> | ||
| + | <li> Fluorescent proteins RFP and sfGFP </li> | ||
| + | <li> 5 chromoproteins </li> | ||
| + | <li> 11 CBD fusion proteins </li> | ||
| + | <li> Vitreoscilla hemoglobin gene </li> | ||
| + | <li> TetR and Arabinose inducible promoters </li> | ||
| + | </ul> | ||
| + | </p> | ||
| + | <p> Because the toolkit is in the non-standard pSEVA331-Bb backbone, it can not be housed in the Parts Registry, however in order to make it accessible for the synthetic biology community, we have made it freely available on request. | ||
| + | </p> | ||
<div class="accordion"> | <div class="accordion"> | ||
| - | <h3> | + | <h3>Characterization of G.xylinus ATCC 53582 and igem strains</h3> |
<div> | <div> | ||
| - | <p> | + | <p> Parts: |
| - | + | -<a href="http://parts.igem.org/Part:BBa_K1321305">BBa_K1321305</a> | |
| - | + | -<a href="http://parts.igem.org/Part:BBa_K1321306">BBa_K1321306</a> | |
| - | + | </p> | |
| + | <p> Cellulose productivity of G.xylinus ATCC 53582 and igem on HS-glucose</p> | ||
| + | <p> G.xylinus cellulose productivity depends strongly on the carbon feedstocks used. In HS-glucose media grown for 7 days at 30degC standing using 50ml HS medium in 250ml conical flasks (sealed with foam buns), cellulose productivity of the igem strain is approximately one fifth of that of ATCC 53582 strain (see Figure 4 for comparison of ATCC53582 and igem strains and experimental details). ATCC 53582 strain produces around 0.5-0.6g of pure cellulose after 5 days of incubation in these conditions. As 50ml HS medium contains 1g of glucose, ATCC 53582 strain converts glucose to cellulose with around 50-60% efficiency. However, it seems for igem cellulose productivity, HS-glucose, is not optimal, because in HS-glycerol media and HS-sucrose media the cellulose productivity is increased. Importantly, in HS-sucrose medium, cellulose productivity is higher than that of ATCC 53582, which is of industrial significance due to the high availability and low cost of sucrose (see Feedstocks for G.xylinus). | ||
| + | <figure class="content-image image-left"> | ||
| + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/b/b3/IC14-ATCC_and_KI_cellulose_productivity_in_HS_correct.jpg"> | ||
| + | <figcaption> Figure 4. Cellulose production of G.xylinus ATCC 53582 and igem (KI) strains in HS-glucose medium. Cultures were inoculated using 50µl of 48h-grown seed culture, and grown in 50ml HS in 250ml conical flasks, sealed with foam buns at 30degC standing. Cellulose was washed twice with dH20, treated with 0.1M NaOH for 4 hours at 60degC, washed twice with dH20 and dried at 60degC for 48h before measuring cellulose pellicle weight. N=3, error bars denote SD. </figcaption> | ||
| + | </figure> | ||
| + | </p> | ||
| + | <p>Cellulose productivity on different carbon feedstocks </p> | ||
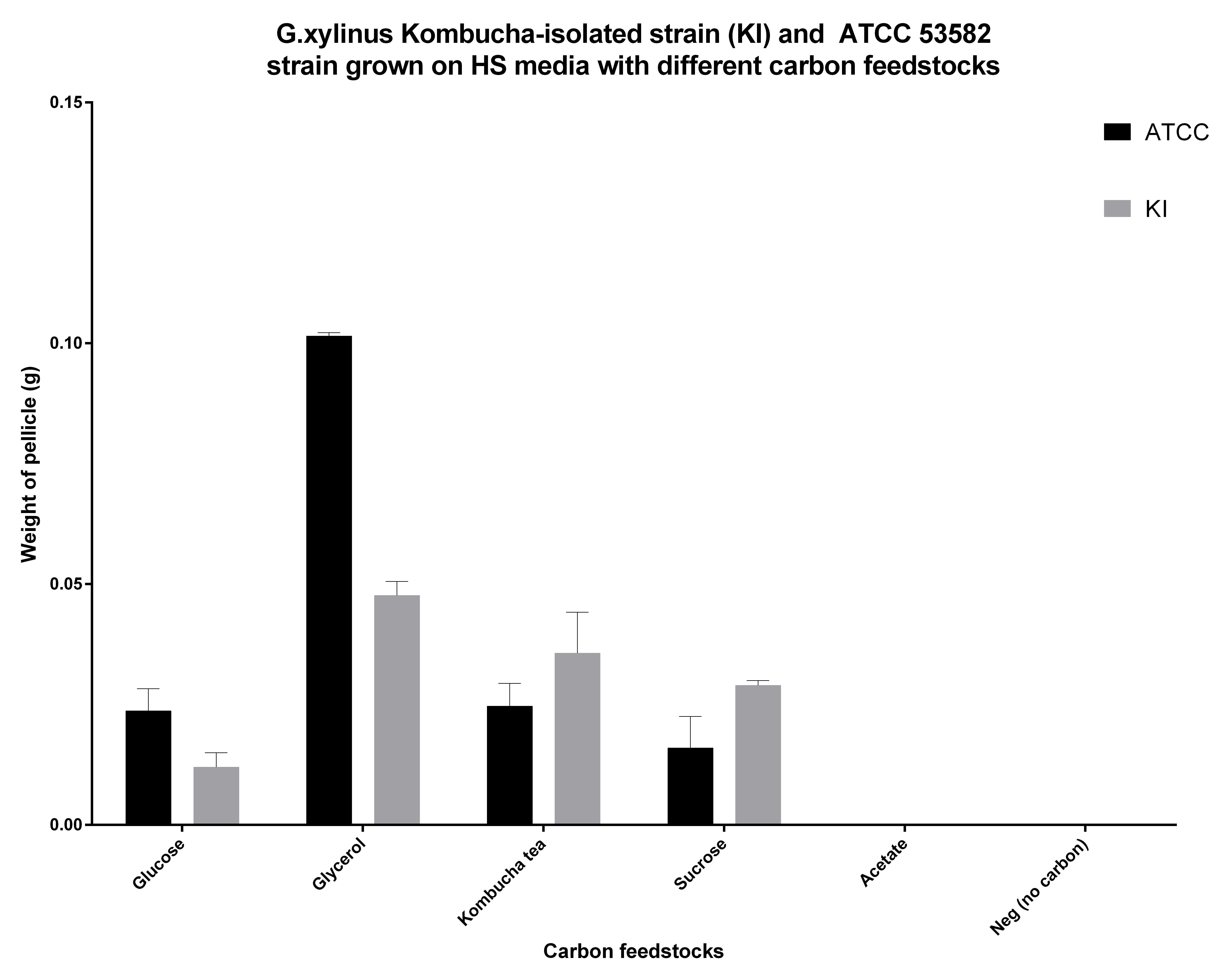
| + | <p>G.xylinus is commonly grown on HS-glucose. However, it can readily use other carbon feedstocks, including sucrose and glycerol. We have found that ATCC 53582 cellulose productivity is highest when grown on glycerol (see Figure 5.), although the effects of routine culturing of ATCC 53582 on HS-glycerol media are unknown. Suprisingly however, igem strain has a higher cellulose productivity in HS-sucrose than G.xylinus ATCC 53582 (see Figure 5 for comparison of ATCC and igem strains and details of he experiment). This could be due to the niche igem strain has evolved into, as the Kombucha tea contains high concentrations (commonly around 100g/L) of sucrose. </p> | ||
| + | <figure class="content-image image-left"> | ||
| + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/5/5c/IC14_ATCC_and_KI_carbon_feedstocks_experimentII_with_acetate_and_negative.jpg"> | ||
| + | <figcaption> Figure 5. Cellulose productivity of G.xylinum ATCC 53582 and igem (KI) strains in HS medium containing different carbon feedstocks. Glycerol seems to result in highest productivity, however the effects of glycerol on routine culture are not known. All carbon feedstocks were normalized to 2% (w/v). Cells were cultured in 50ml Corning tubes, filled with 20ml HS medium standing at 30C for 10 days, cultured with loose caps to allow diffusion of air, and kept at 4degC until measurement. Cellulose was washed twice with dH20, treated with 0.1M NaOH for 4 hours at 60degC, washed twice with dH20 and dried at 60degC for 48h before measuring cellulose pellicle weight. N=3, error bars denote SD.</figcaption> | ||
| + | </figure> | ||
| + | </p> | ||
| + | <p> Natural antibiotics resistance of ATCC 53582 and igem strains </p> | ||
| + | <p></p> | ||
| + | <figure class="content-image image-left"> | ||
| + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/4/4c/IC14_XYZNatural_antibiotics_resistance_of_ATCC_53582_and_KI_strains.jpg"> | ||
| + | <figcaption> Figure 6. Natural antibiotics resistance of G.xylinus ATCC 53582 and igem (KI) strains. Due to small colony diameter and large clustering, colonies were rounded to the nearest hundred, if more than a hundred colonies were present. Plates with over 500 colonies often formed a lawn. Plates were seeded with 50µl of ATCC and igem seed culture normalized to equal OD600, and grown inverted at 30degC for 5 days until colony counting. N=3, error bars denote SD. </figcaption> | ||
| + | </figure> | ||
| + | <p>Using optimal antibiotics concentrations is critical for selection of transformed cells while not inhibiting the cell growth. G.xylinus ATCC 53582 is not resistant to kanamycin and ampicillin in concentrations normally used for E.coli (50µg/ml and 100µg/ml respectively), however is resistant to chloramphenicol (at 35µg/ml; see Figure 6 for an experiment of ATCC 53582 natural antibiotic concentrations, experimental details, and comparison of ATCC and igem strains). However, when using high cell densities for plating (after transformations), antibiotic-resistant colonies of ATCC 53582 appear at up to 3x concentrations. Thus, the recommended antibiotic concentrations for HS-agar plates are 200µg/ml kanamyicn, 400µg/ml ampicillin and 140µg/ml chloramphenicol.</p> | ||
| + | <p>G.xylinus igem is naturally resistant to kanamycin, ampicillin and chloramphenicol concentrations normally used for E.coli. Also, we have found that the frequency of appearance of antibiotic-resistant colonies is dependent on the number of cells used for plating - with higher cell numbers, resistant colonies can be found on higher antibiotics concentrations. We have found that the igem strain forms antibiotic-resistant colonies up to 6x antibiotic concentrations used for E.coli, thus the recommended amounts of antibiotics for transformations are 350µg/ml kanamycin, 700µg/ml ampicillin or 245µg/ml chloramphenicol.</p> | ||
| + | |||
| + | |||
</div> | </div> | ||
| - | <h3> | + | <h3>Plasmid backbones</h3> |
<div> | <div> | ||
| - | + | <figure class="content-image"> | |
| - | + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/3/31/IC14-Plasmid_backbones_in_G.xylinus.jpg"> | |
| - | + | <figcaption>Figure 7. PCR confirmation of replication of pSEVA321, pSEVA331, pSEVA351, pBla and pBAV1K in G.xylinus. DNA source is miniprepped plasmid DNA from G.xylinus igem after transformation and culturing (we had similar results with the ATCC 53582 strain.) L- NEB 2-log ladder, Ctr- Negative control, roman numerals denote replicates. Positive controls were miniprepped from E.coli prior to the experiment. Expected band sizes of positive results: pSEVA321-351: 328bp, pBla approximately 700bp, pBAV1K-985bp. 1% agarose gel, run at 100V, 20 min.</figcaption> | |
| - | + | </figure> | |
| - | + | <p>We tested 9 different plasmid backbones previously not known to replicate in G.xylinus - pSEVA311, pSEVA321, pSEVA331, pSEVA341, pSEVA351, pBla-Vhb-122 (from Chien et al. 2006), pBAV1K-T5-sfgfp, pSB1C3 and pBca1020 (part number BBa_J61002) in order to create a set of plasmid backbones with different copy numbers for G.xylinus genetic engineering. We used pBla-Vhb-122 as the positive control, as it has been previously shown to replicate in G.xylinus (see Chien et al. 2006). In order to test for replication, we prepared two different sets of electrocompetent cells from both ATCC 53582 and igem strains (see <a href="https://2014.igem.org/Team:Imperial/Protocols">Preparing electrocompetent cells</a>) and transformed using 33µl, 50µl and 100µl of electrocompetent cells and 50µg, 200µg and 500µg of DNA (see <a href="https://2014.igem.org/Team:Imperial/Protocols">Transformation of G.xylinus using electroporation</a>). Transformants were plated out on 4 different antibiotic concentrations for each transformation: 0.5x, 1x, 2x and 6x concentrations (this refers to concentrations of antibiotics commonly used for E.coli, which are 50µg/ml kanamycin, 100µg/ml ampicillin and 35µg/ml chloramphenicol). Colonies were then inoculated into HS-cellulase medium and grown for 3 days shaking at 180RPM with appropriate antibiotics, after which plasmid DNA was miniprepped using the QiaPrep Spin Miniprep kit, according to manufacturer's instructions. Miniprepped DNA was then used as a template for PCR to determine plasmid-containing colonies. Primers used for PCR had been previously verified with pure plasmid DNA. DNA sequence of successfully amplified plasmids were confirmed by Sanger sequencing.</p> | |
| - | + | <p>We discovered that 5 out of the 9 tested plasmids - pSEVA321, pSEVA331, pSEVA351, pBAV1K and pBla-Vhb-122 (as the positive control)- were capable of replication in both G.xylinus igem and ATCC 53583 (See Figure 7). No positive results were obtained for other plasmids, indicating that the rest can not replicate in G.xylinus, or were not transformed due to other reasons. We converted pSEVA321 and pSEVA331 into a biobrick-compatible format by inserting the biobrick prefix and suffix in place of the original multiple cloning site using PCR with mutagenic primers and are currently in the process of converting pSEVA351 and pBla-Vhb-122 (converting them into biobrick format has required more time, as they contain several forbidden restriction enzyme sites requiring multiple steps of mutagenesis). pBAV1K was already engineered to be biobrick compatible by authors. We have also verified replication of all of these plasmids in E.coli. Thus, we have determined 5 new plasmid backbones that can act as shuttle vectors for G.xylinus genetic engineering: </p> | |
| - | + | <ul> | |
| - | + | <li> pSEVA321-Bb - part <a href="http://parts.igem.org/Part:BBa_K1321301">BBa_K1321301</a> </li> | |
| - | + | <li> pSEVA331-Bb - part <a href="http://parts.igem.org/Part:BBa_K1321300">BBa_K1321300</a> | |
| - | + | <li> pSEVA351-Bb - part <a href="http://parts.igem.org/Part:BBa_K1321307">BBa_K1321307</a> | |
| - | + | <li> pBla-Vhb-122-Bb - part <a href="http://parts.igem.org/Part:BBa_K1321308">BBa_K1321308</a> | |
| - | + | <li> pBAV1K - part <a href="http://parts.igem.org/Part:BBa_K1321309">BBa_K1321309</a> </li> | |
| - | + | </ul> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | <h3> | + | <h3>Anderson promoters</h3> |
<div> | <div> | ||
| - | + | <p> We inserted 15 Anderson promoter-RFP coding devices into pSEVA331-Bb backbone, transformed them into G.xylinus igem strain in order to determine their relative strengths in Gluconacetobacter (see Figure 8 for some of them: | |
| - | + | <figure class="content-image image-left"> | |
| - | + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/1/1a/IC14_Composite_image_correct.jpg"> | |
| + | <figcaption>Figure 8. Different Anderson promoter-RFP constructs expressed in G.xylinus. Left- wild-type negative control. Right- G.xylinus igem transformed with pSEVA331Bb-Anderson promoter constructs. Plates grown at 30C for 7 days before imageing under blue light. </figcaption> | ||
| + | </figure> | ||
| + | </p> | ||
</div> | </div> | ||
| - | + | <h3>Vitreoscilla hemoglobin</h3> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <h3> | + | |
<div> | <div> | ||
| - | <p> | + | <p><i>G. xylinus</i> is obligatively aerobic because of which cellulose production is reduced below the medium surface in static culture[1] due to reduced oxygen concentrations, reducing the overall volume of the pellicle that is biologically active. Transforming <i>G. xylinus</i> with the gene for bacterial hemoglobin from <i>Vitreoscilla</i> (another obligate aerobe that expresses this protein in oxygen-poor environments) has been shown to result in increased the metabolic activity and cellulose production of G.xylinus [2]. |
| - | + | Vitreoscilla</i> hemoglobin (VHb) is a monomeric heme-containing protein that appears to improve the metabolic function of obligate aerobes and facultative anaerobes in low-oxygen conditions[3][4][5][6]. Evidence suggests that the protein binds oxygen, then shuttles it to at least one cytochrome in the electron transport chain[7], improving the rate of oxidative phosphorylation and therefore ATP production even when dissolved oxygen is scarce.</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <figure class="content-image image-left"> | |
| - | + | <img class="image-full" src="https://static.igem.org/mediawiki/2014/f/f6/IC14_Vhb_effects_on_growth_correct.jpg"> | |
| - | + | <figcaption>Figure 9. Effects of Vitreoscilla hemoglobin expression on G.xylinus maximum biomass production. <em>G.xylinus </em> igem wild type cells and cells transformed with pSEVA331-BBa_K1321200 plasmid were cultured in 5ml of HS-cellulase medium (in 50ml Falcon tubes with loose caps) at 30degC, 180rpm shaking for 4 days, after which OD600 was measured. Samples were diluted 1:1 in HS-cellulase medium before measurement. Negative controls (HS-cellulase without inoculations) showed no growth. N=3 (Vhb) and 4(wild-type), error bars denote SD. </figcaption> | |
| - | + | </figure> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p>We cloned the Vhb gene behind a medium-to-strong Anderson promoter J23101 and the Elowitz RBS (part <a href="http://parts.igem.org/Part:BBa_K1321200">BBa_K1321200</a>) in pSEVA331-Bb backbone in order to increase growth rate and cellulose productivity of <em>G. xylinus</em>. We chose J23101 in order not to overburden the cell with protein expression, while still providing a large amount of Vhb. Expression of Vhb increased the final culture density of <em>G.xylinus</em> igem strain almost two-fold (with a high statistical significance of p<0.0001; see Figure 9) when grown to stationary phase in HS-cellulase medium, demonstrating that expression of Vhb increases the maximum G.xylinus biomass. </p> | |
| - | + | ||
| - | + | <p>We also made a hypothesis of the metabolic pathway involving the interaction of vHb, which could potentially explain its effect on enhancing bacterial cellulose yield. As described in the diagram above, vHb plays a role as oxygen storage: it will actively capture oxygen molecules when when the oxygen concentration is high in the cell whereas release oxygen molecules when it is low. As such, the presence of vHb will smooth out the oxygen level within the cell and therefore provides it with a relatively constant oxygen supply. Considering the fact that oxygen also bind to protein complex cG PDE (cyclic GMP phosphodiesterase), where the binding will impose an upregulating effect on bacterial cellulose production, a constant oxygen level maintained by vHb will inevitably contribute to enhance bacterial cellulose yield.</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <figure class="content-image image-left"> | ||
| + | <img class="image-full" src="https://lh5.googleusercontent.com/OkLBTdjMaO6PjVewhOFchlpEd3x4REDL53KYLp_sSxkD35RU4U8Gh21PJKUSfTA1GZ7ShEayn0Y=w751-h608"> | ||
| + | <figcaption>Figure 10. Effects of vHb interaction on cellular oxygen level.</figcaption> | ||
| + | </figure> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
| - | + | <section id="references"> | |
| + | <h2>References</h2> | ||
| + | <ul> | ||
| - | + | <li>[1] https://www.researchgate.net/ publication/227802179_Optimizing_the_Production_of_Bacterial_Cellulose_in_Surface_Culture_Evaluation_of_Substrate_Mass_Transfer_Influences_on_the_Bioreaction_%28Part_1%29 - Optimizing the production of bacterial cellulose in surface culture: Evaluation of substrate mass transfer influences on the bioreaction</li> | |
| - | + | <li>[2] http://www.ncbi.nlm.nih.gov/pubmed/17868946 - Expressing Vitreoscilla hemoglobin in statically cultured Acetobacter xylinum with reduced O2 tension maximizes bacterial cellulose pellicle production</li> | |
| - | + | <li>[3] http://www.ncbi.nlm.nih.gov/pubmed/2850971 - Cloning, characterisation and expression of the hemoglobin gene from Vitreoscilla in Escherichia coli</li> | |
| - | + | <li>[4] http://www.ncbi.nlm.nih.gov/pubmed/11478898 - Monomer-dimer equilibrium and oxygen-binding properties of ferrous Vitreoscilla hemoglobin</li> | |
| - | + | <li>[5] http://onlinelibrary.wiley.com/doi/10.1021/bp960071v/full - Expression of Vitreoscilla hemoglobin is superior to horse heart myoglobin or yeast flavohemoglobin for enhancing Escherichia coli growth in a microaerobic bioreactor</li> | |
| - | + | <li>[6] http://www.nature.com/nbt/journal/v11/n8/full/nbt0893-926.html - The production of cephalosporin C by Aecremonium chrysogenum is improved by the intracellular expression of bacterial hemoglobin</li> | |
| - | + | <li>[7] http://onlinelibrary.wiley.com/doi/10.1111/j.1432-1033.1994.tb19931.x/full - Intracellular expression of Vitreoscilla hemoglobin alters Escherichia coli energy metabolism under oxygen-limited conditions</li> | |
| - | + | ||
| - | + | <ul> | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
</section> | </section> | ||
</div> | </div> | ||
Latest revision as of 16:05, 2 February 2015
G. xylinus
Overview
Bacterial cellulose has great potential in many areas, including water purification, tissue scaffolds, wound dressings, etc., however, until now, all bacterial cellulose-based materials have been created using chemical or physical post-production processing, not genetic engineering. This is due to the lack of well-developed tools and methods for Gluconacetobacter genetic engineering, as well as the lack of genome sequence of the highest cellulose-producing strain ATCC 53582. We have overcome the numerous difficulties associated with G.xylinus genetic engineering, and turned G.xylinus igem and ATCC 53852 strains into new platforms for the production of cellulose-based biomaterials by sequencing the genomes of ATCC 53582 and a new strain G. xylinus igem, creating a genetic toolbox of consisting of five new plasmid backbones and around 40 widely used genes, and developing a set of new and improved protocols for G.xylinus genetic engineering.
Key Achievements
- Isolated a new strain of Gluconacetobacter (named G. xylinus igem) from Kombucha tea and characterized its properties fully.
- Sequenced the previously unknown genomes of G. xylinus ATCC 53582 and G. xylinus igem strains - the first genomes sequenced in the history of iGEM
- Discovered four new plasmids capable of replication in Gluconacetobacter species - pSEVA321, pSEVA331, pSEVA351 and pBAV1K, which replicate both in G. xylinus ATCC 53582 and igem strains as well as in E. coli.
- Were the first in science to create transgenic cells of G.xylinus igem strain.
- Using our discovered plasmids, created a genetic toolbox consisting of 40 genes for G. xylinus engineering and expressed them in the ATCC 53582 and igem.
- Developed a set of new and improved protocols for efficient genetic engineering of G. xylinus.
- In summary, turned G. xylinus ATCC 53582 and igem strains into new model organisms and developed the necessary tools to create a platform for genetic engineering of new cellulose-based biomaterials.

Introduction
Due to its high productivity, G. xylinus is the main species used for the production of bacterial cellulose based products. These range from superior wound dressings, artificial blood vessels, scaffolds for tissue engineering, high-quality speaker membranes, stronger paper, nata de coco and many others (Keshk 2014) .
G. xylinus has been the subject of the majority of studies into production of bacterial cellulose. However, research into G. xylinus has primarily focused on the effects of different culture conditions (such as composition of growth media, aeration and agitation) on cellulose productivity, productivity of different strains and different post-processing methods. Few attempts have been made to improve cellulose productivity or the physical properties of cellulose through genetic engineering (but see Chien 2006). Consequently most of the genetic engineering methods and tools available for model organisms, such as E.coli, have not been developed for G. xylinus. These tools must first be developed, in order to begin serious efforts to genetically engineer G. xylinus strains capable of producing novel biomaterials, more cellulose, and at a lower cost. In the middle of the summer, we isolated a new strain of Gluconacetobacter (named G. xylinus igem, which we previously called G. xylinus KI for Kombucha isolated ) from Kombucha tea and characterized its properties fully. We found it to be more amenable for genetic engineering due to higher transformation efficiencies than G. xylinus ATCC 53582 strain (see below), so we continued work on this strain, including sequencing its genome due to potential economic importance (it is a key component in the popular Kombucha tea).
Aims
In addition to the lack of tools, the continuous cellulose production of G. xylinus introduces further problems for genetic engineering, as it results in a low growth rate (the division time of G. xylinus is 4 hours, which is 8 times slower than that of E. coli), formation of spontaneous cellulose non-producing mutants in agitated culture (detrimental for the engineering of high-producing strains) and difficulties in performing procedures such as transformation, plasmid DNA extraction, etc. due to the physically interfering cellulose pellicle.
Furthermore, although the highest cellulose-producing strain G. xylinus ATCC 53582 has been used in several studies, the genome sequence of this strain is still unknown, making it impossible to carry out targeted engineering of chromosomal genes, which is vital to achieve increased productivity.
We aim to solve all of these problems, by completing three major projects: sequencing the genomes of ATCC53582 and igem strains and creating a large toolbox for G. xylinus genetic. We then aim to use these tools to increase and control cellulose productivity and create new biomaterials with wide-ranging properties, by incorporating proteins with different functions into the cellulose matrix (see Functionalisation).
Genome sequencing

Although genome sequencing has become widely available due to the improvements in next-generation sequencing, even with smaller bacterial genomes (the genome of G. xylinus is approximately 3.5Mbp in size), genome sequencing is still unavailable for small to mid-sized research groups due to high costs. We performed a survey of prices offered by all commercially available service providers using the Illumina MiSeq next-generation sequencing platform (which is the most cost-effective method for bacterial genome sequencing) in Europe, and found that the average costs of fully sequencing a 3.5Mbp genome (which includes library preparation, sequencing, bioinformatics and gap filling) is approximately £3000 for "simple" to sequence genomes, but can far exceed that for repeat-rich, GC-rich or otherwise problematic genomes. In order to be able to sequence the genome with the limited budget available to our team, we did not use a commercial sequencing service, and decided to perform the full sequencing cycle ourselves, despite having no previous experience in genome sequencing, and were able to sequence two genomes for less than £1000. We created the gDNA library using Illumina Nextera kit, by modifying the original Illumina protocol to be amenable for small sample number (see gDNA library preparation protocol). During gDNA library preparation, we discovered that cellulose production of G.xylinus resulted in the presence of a persistent contaminant in DNA preparation, which persisted even after four consecutive DNA purification methods. This contamination interfered with effective library preparation, retarding the project several weeks, and ultimately prompted us to create a new and specific protocol for G.xylinus DNA extraction (see G.xylinus DNA extraction). We performed the sequencing run (multiplexed sequencing of ATCC 53582 and igem in a single run, paired-end 250bp reads using standard flow cell) on an Illumina MiSeq at Imperial College’s Genomics Laboratory. Despite problems with gDNA library preparation, the sequencing yielded good results (9,063,619 reads with 98.71% reads identified; see Figure 1 for an example output of read alignment against Komagateibacter xylinus E25 reference genome). We performed the quality control of the reads using FasQC and Trim Galore and completed the genome assembly with the assistance of Imperial Bioinformatics Support service using their in-house assembly pipeline BugBuilder.
As a result, we are proud to present here the first genomes sequenced in iGEM. The genome of G. xylinus igem strain is approximately 3.4Mbp in size, with a GC content of 63.52% (see Figure 2). Autoannotation using Prokka (Seemann 2014) identified in total 3052 putative coding sequences, 59 tRNA sequences and 24 rRNA and other RNA sequences. The genome G. xylinus ATCC53582 is approximately 2.8Mbp in size, with a GC content of 59.82% (see Figure 3). Autoannotation using Prokka (Seemann 2014) identified in total 2491 putative coding sequences, 46 tRNA sequences and 17 rRNA and other RNA sequences. We are currently performing on the comparitive genomics of these genomes and related genomes, and will properly publish our results in a peer-reviewed journal. The full genome sequences, as well as detailed information about G. xylinus igem strain and genome can be found on the Registry as part BBa_K1321306
and G. xylinus ATCC53582 as part BBa_K1321305


Creating a genetic toolbox for G. xylinus
Despite the great potential G. xylinus holds for new biomaterial production, very few tools for genetic engineering had been developed so far. Thus, in parallel to sequencing the genomes, we began building a new system in order to equip G.xylinus with all of the necessary tools and to turn the ATCC 53582 and igem strains into systems for genetically engineered biomaterials production. We first characterized ATCC 53582 and igem strains, and created a set of new and improved protocols necessary for their genetic engineering (see G.xylinus protocols). As only a few vectors are available for G. xylinus genetic engineering (this is due to various reasons including patents, materials-transfer agreements, and retired or deceased researchers) we first created a set of new biobrick-compatible plasmid backbones capable of replication both in Gluconacetobacter and E.coli. In order to create the genetic toolkit, we decided to test out and modify already existing Biobricks in the parts registry (rather than creating a set of new parts) in order to retain the already existing familiarity genetic engineers have with these genes and make the toolkit more user-friendly. None of these parts had been used in Gluconacetobacter before however, so it was necessary to determine if they can be used in G.xylinus in the first place. Finally, we used this toolkit to clone Vitreoscilla hemoglobin gene Vhb (which is known to increase cellulose productivity and growth rate of G.xylinus (Chien et al. 2006)) behind a strong Anderson promoter, and express it in G.xylinus ATCC 53582 and G.xylinus igem strains to increase cellulose productivity and decrease its cost. In total, by cloning widely used parts into our G. xylinus shuttle vectors we created a toolkit consisting of:
- 5 new shuttle vectors
- 15 Anderson promoters
- Fluorescent proteins RFP and sfGFP
- 5 chromoproteins
- 11 CBD fusion proteins
- Vitreoscilla hemoglobin gene
- TetR and Arabinose inducible promoters
Because the toolkit is in the non-standard pSEVA331-Bb backbone, it can not be housed in the Parts Registry, however in order to make it accessible for the synthetic biology community, we have made it freely available on request.
Characterization of G.xylinus ATCC 53582 and igem strains
Parts: -BBa_K1321305 -BBa_K1321306
Cellulose productivity of G.xylinus ATCC 53582 and igem on HS-glucose
G.xylinus cellulose productivity depends strongly on the carbon feedstocks used. In HS-glucose media grown for 7 days at 30degC standing using 50ml HS medium in 250ml conical flasks (sealed with foam buns), cellulose productivity of the igem strain is approximately one fifth of that of ATCC 53582 strain (see Figure 4 for comparison of ATCC53582 and igem strains and experimental details). ATCC 53582 strain produces around 0.5-0.6g of pure cellulose after 5 days of incubation in these conditions. As 50ml HS medium contains 1g of glucose, ATCC 53582 strain converts glucose to cellulose with around 50-60% efficiency. However, it seems for igem cellulose productivity, HS-glucose, is not optimal, because in HS-glycerol media and HS-sucrose media the cellulose productivity is increased. Importantly, in HS-sucrose medium, cellulose productivity is higher than that of ATCC 53582, which is of industrial significance due to the high availability and low cost of sucrose (see Feedstocks for G.xylinus).

Cellulose productivity on different carbon feedstocks
G.xylinus is commonly grown on HS-glucose. However, it can readily use other carbon feedstocks, including sucrose and glycerol. We have found that ATCC 53582 cellulose productivity is highest when grown on glycerol (see Figure 5.), although the effects of routine culturing of ATCC 53582 on HS-glycerol media are unknown. Suprisingly however, igem strain has a higher cellulose productivity in HS-sucrose than G.xylinus ATCC 53582 (see Figure 5 for comparison of ATCC and igem strains and details of he experiment). This could be due to the niche igem strain has evolved into, as the Kombucha tea contains high concentrations (commonly around 100g/L) of sucrose.
Natural antibiotics resistance of ATCC 53582 and igem strains

Using optimal antibiotics concentrations is critical for selection of transformed cells while not inhibiting the cell growth. G.xylinus ATCC 53582 is not resistant to kanamycin and ampicillin in concentrations normally used for E.coli (50µg/ml and 100µg/ml respectively), however is resistant to chloramphenicol (at 35µg/ml; see Figure 6 for an experiment of ATCC 53582 natural antibiotic concentrations, experimental details, and comparison of ATCC and igem strains). However, when using high cell densities for plating (after transformations), antibiotic-resistant colonies of ATCC 53582 appear at up to 3x concentrations. Thus, the recommended antibiotic concentrations for HS-agar plates are 200µg/ml kanamyicn, 400µg/ml ampicillin and 140µg/ml chloramphenicol.
G.xylinus igem is naturally resistant to kanamycin, ampicillin and chloramphenicol concentrations normally used for E.coli. Also, we have found that the frequency of appearance of antibiotic-resistant colonies is dependent on the number of cells used for plating - with higher cell numbers, resistant colonies can be found on higher antibiotics concentrations. We have found that the igem strain forms antibiotic-resistant colonies up to 6x antibiotic concentrations used for E.coli, thus the recommended amounts of antibiotics for transformations are 350µg/ml kanamycin, 700µg/ml ampicillin or 245µg/ml chloramphenicol.
Plasmid backbones
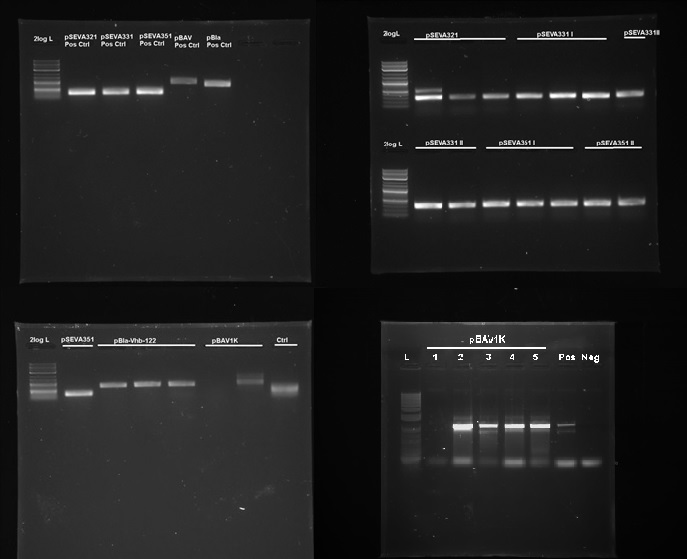
We tested 9 different plasmid backbones previously not known to replicate in G.xylinus - pSEVA311, pSEVA321, pSEVA331, pSEVA341, pSEVA351, pBla-Vhb-122 (from Chien et al. 2006), pBAV1K-T5-sfgfp, pSB1C3 and pBca1020 (part number BBa_J61002) in order to create a set of plasmid backbones with different copy numbers for G.xylinus genetic engineering. We used pBla-Vhb-122 as the positive control, as it has been previously shown to replicate in G.xylinus (see Chien et al. 2006). In order to test for replication, we prepared two different sets of electrocompetent cells from both ATCC 53582 and igem strains (see Preparing electrocompetent cells) and transformed using 33µl, 50µl and 100µl of electrocompetent cells and 50µg, 200µg and 500µg of DNA (see Transformation of G.xylinus using electroporation). Transformants were plated out on 4 different antibiotic concentrations for each transformation: 0.5x, 1x, 2x and 6x concentrations (this refers to concentrations of antibiotics commonly used for E.coli, which are 50µg/ml kanamycin, 100µg/ml ampicillin and 35µg/ml chloramphenicol). Colonies were then inoculated into HS-cellulase medium and grown for 3 days shaking at 180RPM with appropriate antibiotics, after which plasmid DNA was miniprepped using the QiaPrep Spin Miniprep kit, according to manufacturer's instructions. Miniprepped DNA was then used as a template for PCR to determine plasmid-containing colonies. Primers used for PCR had been previously verified with pure plasmid DNA. DNA sequence of successfully amplified plasmids were confirmed by Sanger sequencing.
We discovered that 5 out of the 9 tested plasmids - pSEVA321, pSEVA331, pSEVA351, pBAV1K and pBla-Vhb-122 (as the positive control)- were capable of replication in both G.xylinus igem and ATCC 53583 (See Figure 7). No positive results were obtained for other plasmids, indicating that the rest can not replicate in G.xylinus, or were not transformed due to other reasons. We converted pSEVA321 and pSEVA331 into a biobrick-compatible format by inserting the biobrick prefix and suffix in place of the original multiple cloning site using PCR with mutagenic primers and are currently in the process of converting pSEVA351 and pBla-Vhb-122 (converting them into biobrick format has required more time, as they contain several forbidden restriction enzyme sites requiring multiple steps of mutagenesis). pBAV1K was already engineered to be biobrick compatible by authors. We have also verified replication of all of these plasmids in E.coli. Thus, we have determined 5 new plasmid backbones that can act as shuttle vectors for G.xylinus genetic engineering:
- pSEVA321-Bb - part BBa_K1321301
- pSEVA331-Bb - part BBa_K1321300
- pSEVA351-Bb - part BBa_K1321307
- pBla-Vhb-122-Bb - part BBa_K1321308
- pBAV1K - part BBa_K1321309
Anderson promoters
We inserted 15 Anderson promoter-RFP coding devices into pSEVA331-Bb backbone, transformed them into G.xylinus igem strain in order to determine their relative strengths in Gluconacetobacter (see Figure 8 for some of them:

Vitreoscilla hemoglobin
G. xylinus is obligatively aerobic because of which cellulose production is reduced below the medium surface in static culture[1] due to reduced oxygen concentrations, reducing the overall volume of the pellicle that is biologically active. Transforming G. xylinus with the gene for bacterial hemoglobin from Vitreoscilla (another obligate aerobe that expresses this protein in oxygen-poor environments) has been shown to result in increased the metabolic activity and cellulose production of G.xylinus [2]. Vitreoscilla hemoglobin (VHb) is a monomeric heme-containing protein that appears to improve the metabolic function of obligate aerobes and facultative anaerobes in low-oxygen conditions[3][4][5][6]. Evidence suggests that the protein binds oxygen, then shuttles it to at least one cytochrome in the electron transport chain[7], improving the rate of oxidative phosphorylation and therefore ATP production even when dissolved oxygen is scarce.

We cloned the Vhb gene behind a medium-to-strong Anderson promoter J23101 and the Elowitz RBS (part BBa_K1321200) in pSEVA331-Bb backbone in order to increase growth rate and cellulose productivity of G. xylinus. We chose J23101 in order not to overburden the cell with protein expression, while still providing a large amount of Vhb. Expression of Vhb increased the final culture density of G.xylinus igem strain almost two-fold (with a high statistical significance of p<0.0001; see Figure 9) when grown to stationary phase in HS-cellulase medium, demonstrating that expression of Vhb increases the maximum G.xylinus biomass.
We also made a hypothesis of the metabolic pathway involving the interaction of vHb, which could potentially explain its effect on enhancing bacterial cellulose yield. As described in the diagram above, vHb plays a role as oxygen storage: it will actively capture oxygen molecules when when the oxygen concentration is high in the cell whereas release oxygen molecules when it is low. As such, the presence of vHb will smooth out the oxygen level within the cell and therefore provides it with a relatively constant oxygen supply. Considering the fact that oxygen also bind to protein complex cG PDE (cyclic GMP phosphodiesterase), where the binding will impose an upregulating effect on bacterial cellulose production, a constant oxygen level maintained by vHb will inevitably contribute to enhance bacterial cellulose yield.
References
- [1] https://www.researchgate.net/ publication/227802179_Optimizing_the_Production_of_Bacterial_Cellulose_in_Surface_Culture_Evaluation_of_Substrate_Mass_Transfer_Influences_on_the_Bioreaction_%28Part_1%29 - Optimizing the production of bacterial cellulose in surface culture: Evaluation of substrate mass transfer influences on the bioreaction
- [2] http://www.ncbi.nlm.nih.gov/pubmed/17868946 - Expressing Vitreoscilla hemoglobin in statically cultured Acetobacter xylinum with reduced O2 tension maximizes bacterial cellulose pellicle production
- [3] http://www.ncbi.nlm.nih.gov/pubmed/2850971 - Cloning, characterisation and expression of the hemoglobin gene from Vitreoscilla in Escherichia coli
- [4] http://www.ncbi.nlm.nih.gov/pubmed/11478898 - Monomer-dimer equilibrium and oxygen-binding properties of ferrous Vitreoscilla hemoglobin
- [5] http://onlinelibrary.wiley.com/doi/10.1021/bp960071v/full - Expression of Vitreoscilla hemoglobin is superior to horse heart myoglobin or yeast flavohemoglobin for enhancing Escherichia coli growth in a microaerobic bioreactor
- [6] http://www.nature.com/nbt/journal/v11/n8/full/nbt0893-926.html - The production of cephalosporin C by Aecremonium chrysogenum is improved by the intracellular expression of bacterial hemoglobin
- [7] http://onlinelibrary.wiley.com/doi/10.1111/j.1432-1033.1994.tb19931.x/full - Intracellular expression of Vitreoscilla hemoglobin alters Escherichia coli energy metabolism under oxygen-limited conditions
 "
"











